Please be patient as the page loads
|
MMP9_MOUSE
|
||||||
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MMP9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990348 (rank : 2) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 151 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP2_HUMAN
|
||||||
| θ value | 1.79534e-149 (rank : 3) | NC score | 0.972531 (rank : 3) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| θ value | 2.59246e-148 (rank : 4) | NC score | 0.972362 (rank : 4) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP3_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 5) | NC score | 0.862444 (rank : 9) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP13_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 6) | NC score | 0.864444 (rank : 7) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP10_MOUSE
|
||||||
| θ value | 8.59492e-43 (rank : 7) | NC score | 0.862162 (rank : 10) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
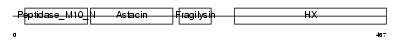
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP10_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 8) | NC score | 0.861870 (rank : 11) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP13_MOUSE
|
||||||
| θ value | 5.57106e-42 (rank : 9) | NC score | 0.863748 (rank : 8) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP3_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 10) | NC score | 0.861762 (rank : 12) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP11_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 11) | NC score | 0.856391 (rank : 20) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP1_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 12) | NC score | 0.861101 (rank : 13) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP8_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 13) | NC score | 0.858026 (rank : 19) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP1A_MOUSE
|
||||||
| θ value | 5.96599e-36 (rank : 14) | NC score | 0.858241 (rank : 16) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | 7.79185e-36 (rank : 15) | NC score | 0.855882 (rank : 22) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 16) | NC score | 0.259987 (rank : 53) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 17) | NC score | 0.259449 (rank : 54) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
MMP12_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 18) | NC score | 0.856265 (rank : 21) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP20_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 19) | NC score | 0.858041 (rank : 18) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP12_MOUSE
|
||||||
| θ value | 5.584e-34 (rank : 20) | NC score | 0.858692 (rank : 15) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP20_MOUSE
|
||||||
| θ value | 2.12192e-33 (rank : 21) | NC score | 0.858077 (rank : 17) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP1B_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 22) | NC score | 0.855415 (rank : 23) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP7_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 23) | NC score | 0.870127 (rank : 5) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |
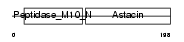
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP8_HUMAN
|
||||||
| θ value | 3.3877e-31 (rank : 24) | NC score | 0.855403 (rank : 24) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP7_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 25) | NC score | 0.869868 (rank : 6) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |
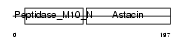
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP14_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 26) | NC score | 0.838981 (rank : 29) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 27) | NC score | 0.838313 (rank : 30) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP15_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 28) | NC score | 0.832942 (rank : 35) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP24_MOUSE
|
||||||
| θ value | 7.06379e-29 (rank : 29) | NC score | 0.839026 (rank : 28) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP16_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 30) | NC score | 0.835232 (rank : 32) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP24_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 31) | NC score | 0.837171 (rank : 31) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP17_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 32) | NC score | 0.840960 (rank : 26) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP16_MOUSE
|
||||||
| θ value | 2.68423e-28 (rank : 33) | NC score | 0.834995 (rank : 33) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP15_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 34) | NC score | 0.829130 (rank : 37) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP17_MOUSE
|
||||||
| θ value | 7.80994e-28 (rank : 35) | NC score | 0.840859 (rank : 27) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | 9.54697e-26 (rank : 36) | NC score | 0.842668 (rank : 25) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP19_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 37) | NC score | 0.833298 (rank : 34) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |
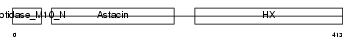
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP26_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 38) | NC score | 0.860856 (rank : 14) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP28_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 39) | NC score | 0.831066 (rank : 36) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP19_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 40) | NC score | 0.827634 (rank : 38) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 41) | NC score | 0.394510 (rank : 46) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 42) | NC score | 0.399106 (rank : 45) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 43) | NC score | 0.265533 (rank : 52) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 44) | NC score | 0.271068 (rank : 51) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 45) | NC score | 0.255808 (rank : 56) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 46) | NC score | 0.256119 (rank : 55) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MMP21_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 47) | NC score | 0.778044 (rank : 40) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MPRI_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 48) | NC score | 0.290683 (rank : 48) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
MMP21_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 49) | NC score | 0.784377 (rank : 39) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MPRI_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 50) | NC score | 0.284525 (rank : 49) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
FA12_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 51) | NC score | 0.095197 (rank : 59) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 52) | NC score | 0.092726 (rank : 60) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
LY75_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 53) | NC score | 0.240768 (rank : 57) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
HGFA_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 54) | NC score | 0.091919 (rank : 62) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
LY75_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 55) | NC score | 0.236931 (rank : 58) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 56) | NC score | 0.043708 (rank : 78) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 57) | NC score | 0.039356 (rank : 80) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 58) | NC score | 0.082733 (rank : 63) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
HEMO_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 59) | NC score | 0.509711 (rank : 44) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |
|
|||||
| Description | Hemopexin precursor. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 60) | NC score | 0.044238 (rank : 77) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 61) | NC score | 0.032402 (rank : 88) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 62) | NC score | 0.026083 (rank : 100) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 63) | NC score | 0.588614 (rank : 41) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
SPRR3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 64) | NC score | 0.076741 (rank : 64) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 65) | NC score | 0.560333 (rank : 42) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 66) | NC score | 0.027580 (rank : 96) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CP003_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 67) | NC score | 0.092672 (rank : 61) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
KPCD3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 68) | NC score | 0.000313 (rank : 151) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94806 | Gene names | PRKD3, EPK2, PRKCN | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D3 (EC 2.7.11.13) (Protein kinase C nu type) (nPKC-nu) (Protein kinase EPK2). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 69) | NC score | 0.046271 (rank : 75) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 70) | NC score | 0.029263 (rank : 92) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
KPCD3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 71) | NC score | 0.000043 (rank : 154) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1Y2 | Gene names | Prkd3, Prkcn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase D3 (EC 2.7.11.13) (Protein kinase C nu type) (nPKC-nu). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 72) | NC score | 0.012212 (rank : 128) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 73) | NC score | 0.019402 (rank : 109) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 74) | NC score | 0.019963 (rank : 107) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 75) | NC score | 0.040901 (rank : 79) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 76) | NC score | 0.028620 (rank : 93) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 77) | NC score | 0.026600 (rank : 98) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 78) | NC score | 0.035728 (rank : 82) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
TSNA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 79) | NC score | 0.035416 (rank : 83) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 0.21417 (rank : 80) | NC score | 0.021540 (rank : 106) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 0.279714 (rank : 81) | NC score | 0.052661 (rank : 70) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 0.279714 (rank : 82) | NC score | 0.052035 (rank : 71) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
K0240_MOUSE
|
||||||
| θ value | 0.279714 (rank : 83) | NC score | 0.023427 (rank : 102) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 84) | NC score | 0.329388 (rank : 47) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 85) | NC score | 0.018565 (rank : 112) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 86) | NC score | 0.000157 (rank : 152) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 87) | NC score | 0.000802 (rank : 150) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.47712 (rank : 88) | NC score | 0.035010 (rank : 85) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 89) | NC score | 0.034093 (rank : 86) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 90) | NC score | 0.031751 (rank : 89) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 91) | NC score | 0.028516 (rank : 94) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DCX_HUMAN
|
||||||
| θ value | 0.62314 (rank : 92) | NC score | 0.015262 (rank : 120) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
GLI1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 93) | NC score | -0.000709 (rank : 155) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 94) | NC score | 0.012554 (rank : 126) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
ATS13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 95) | NC score | 0.062115 (rank : 67) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 96) | NC score | 0.035834 (rank : 81) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 97) | NC score | 0.035294 (rank : 84) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 98) | NC score | 0.008309 (rank : 139) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
ATS4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.013448 (rank : 122) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BNJ2, Q8K384 | Gene names | Adamts4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-4 precursor (EC 3.4.24.82) (A disintegrin and metalloproteinase with thrombospondin motifs 4) (ADAM-TS 4) (ADAM-TS4) (Aggrecanase-1). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 100) | NC score | 0.021558 (rank : 104) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
FOXO4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 101) | NC score | 0.017774 (rank : 114) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
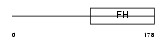
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
HEMO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 102) | NC score | 0.550910 (rank : 43) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |
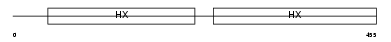
|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 103) | NC score | 0.012164 (rank : 129) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 104) | NC score | 0.033193 (rank : 87) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ATS4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | 0.013270 (rank : 123) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75173, Q5VTW2, Q6P4Q8, Q6UWA8, Q9UN83 | Gene names | ADAMTS4, KIAA0688 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-4 precursor (EC 3.4.24.82) (A disintegrin and metalloproteinase with thrombospondin motifs 4) (ADAM-TS 4) (ADAM-TS4) (Aggrecanase-1) (ADMP-1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 106) | NC score | 0.013202 (rank : 124) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.38821 (rank : 107) | NC score | 0.027876 (rank : 95) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.008660 (rank : 137) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.026463 (rank : 99) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 110) | NC score | 0.011952 (rank : 130) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.019580 (rank : 108) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 112) | NC score | 0.016667 (rank : 115) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 113) | NC score | 0.029641 (rank : 90) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 114) | NC score | 0.009256 (rank : 136) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
3BHS6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.015317 (rank : 119) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35469 | Gene names | Hsd3b6 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type VI (3Beta-HSD VI) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.002650 (rank : 147) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.011233 (rank : 132) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | -0.003911 (rank : 159) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | -0.001476 (rank : 157) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.012533 (rank : 127) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.012629 (rank : 125) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.010841 (rank : 133) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
FREA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.006303 (rank : 144) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.006518 (rank : 143) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
DOS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.015868 (rank : 117) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
EPOR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.013821 (rank : 121) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19235, Q15443 | Gene names | EPOR | |||
|
Domain Architecture |
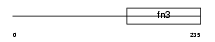
|
|||||
| Description | Erythropoietin receptor precursor (EPO-R). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.004575 (rank : 145) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.011666 (rank : 131) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.025093 (rank : 101) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.000085 (rank : 153) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
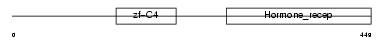
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
ATS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.021779 (rank : 103) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.021547 (rank : 105) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.019324 (rank : 110) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15072, Q9BXZ8 | Gene names | ADAMTS3, KIAA0366 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-3 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 3) (ADAM-TS 3) (ADAM-TS3) (Procollagen II amino propeptide-processing enzyme) (Procollagen II N-proteinase) (PC II-NP). | |||||
|
CRRY_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.002075 (rank : 149) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.007517 (rank : 142) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
FTS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.007579 (rank : 141) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8T0 | Gene names | FTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein homolog (Ft1). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.018364 (rank : 113) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.018648 (rank : 111) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.026675 (rank : 97) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
ZBT45_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | -0.002287 (rank : 158) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.008297 (rank : 140) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ADA15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.008452 (rank : 138) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O88839, Q3U7C2, Q8C7Z0, Q91VS9, Q9QYL2 | Gene names | Adam15, Mdc15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin) (AD56). | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.016315 (rank : 116) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.029507 (rank : 91) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
C1QR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.010403 (rank : 134) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.015443 (rank : 118) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
DCX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.009896 (rank : 135) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.002545 (rank : 148) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | -0.000889 (rank : 156) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SELV_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.045559 (rank : 76) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
VASN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.004433 (rank : 146) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZT5, Q8BJJ0, Q8R2G5 | Gene names | Vasn, Slitl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
CD302_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.070693 (rank : 65) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.069800 (rank : 66) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CHODL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.056807 (rank : 68) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.056647 (rank : 69) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
EMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.050333 (rank : 74) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
PRG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.051512 (rank : 72) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.271796 (rank : 50) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.051358 (rank : 73) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 151 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.990348 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP2_HUMAN
|
||||||
| NC score | 0.972531 (rank : 3) | θ value | 1.79534e-149 (rank : 3) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| NC score | 0.972362 (rank : 4) | θ value | 2.59246e-148 (rank : 4) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MMP7_HUMAN
|
||||||
| NC score | 0.870127 (rank : 5) | θ value | 4.72714e-33 (rank : 23) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| NC score | 0.869868 (rank : 6) | θ value | 9.8567e-31 (rank : 25) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.864444 (rank : 7) | θ value | 3.85808e-43 (rank : 6) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP13_MOUSE
|
||||||
| NC score | 0.863748 (rank : 8) | θ value | 5.57106e-42 (rank : 9) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP3_HUMAN
|
||||||
| NC score | 0.862444 (rank : 9) | θ value | 5.95217e-44 (rank : 5) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.862162 (rank : 10) | θ value | 8.59492e-43 (rank : 7) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP10_HUMAN
|
||||||
| NC score | 0.861870 (rank : 11) | θ value | 2.50074e-42 (rank : 8) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP3_MOUSE
|
||||||
| NC score | 0.861762 (rank : 12) | θ value | 1.05066e-40 (rank : 10) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP1_HUMAN
|
||||||
| NC score | 0.861101 (rank : 13) | θ value | 1.85459e-37 (rank : 12) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP26_HUMAN
|
||||||
| NC score | 0.860856 (rank : 14) | θ value | 5.79196e-23 (rank : 38) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP12_MOUSE
|
||||||
| NC score | 0.858692 (rank : 15) | θ value | 5.584e-34 (rank : 20) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP1A_MOUSE
|
||||||
| NC score | 0.858241 (rank : 16) | θ value | 5.96599e-36 (rank : 14) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP20_MOUSE
|
||||||
| NC score | 0.858077 (rank : 17) | θ value | 2.12192e-33 (rank : 21) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_HUMAN
|
||||||
| NC score | 0.858041 (rank : 18) | θ value | 3.27365e-34 (rank : 19) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP8_MOUSE
|
||||||
| NC score | 0.858026 (rank : 19) | θ value | 4.568e-36 (rank : 13) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.856391 (rank : 20) | θ value | 1.16164e-39 (rank : 11) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP12_HUMAN
|
||||||
| NC score | 0.856265 (rank : 21) | θ value | 1.9192e-34 (rank : 18) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.855882 (rank : 22) | θ value | 7.79185e-36 (rank : 15) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP1B_MOUSE
|
||||||
| NC score | 0.855415 (rank : 23) | θ value | 2.77131e-33 (rank : 22) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP8_HUMAN
|
||||||
| NC score | 0.855403 (rank : 24) | θ value | 3.3877e-31 (rank : 24) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.842668 (rank : 25) | θ value | 9.54697e-26 (rank : 36) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP17_HUMAN
|
||||||
| NC score | 0.840960 (rank : 26) | θ value | 2.05525e-28 (rank : 32) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP17_MOUSE
|
||||||
| NC score | 0.840859 (rank : 27) | θ value | 7.80994e-28 (rank : 35) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| NC score | 0.839026 (rank : 28) | θ value | 7.06379e-29 (rank : 29) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP14_HUMAN
|
||||||
| NC score | 0.838981 (rank : 29) | θ value | 4.89182e-30 (rank : 26) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| NC score | 0.838313 (rank : 30) | θ value | 3.17079e-29 (rank : 27) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP24_HUMAN
|
||||||
| NC score | 0.837171 (rank : 31) | θ value | 1.57365e-28 (rank : 31) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP16_HUMAN
|
||||||
| NC score | 0.835232 (rank : 32) | θ value | 1.2049e-28 (rank : 30) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| NC score | 0.834995 (rank : 33) | θ value | 2.68423e-28 (rank : 33) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP19_MOUSE
|
||||||
| NC score | 0.833298 (rank : 34) | θ value | 3.39556e-23 (rank : 37) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP15_HUMAN
|
||||||
| NC score | 0.832942 (rank : 35) | θ value | 7.06379e-29 (rank : 28) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP28_HUMAN
|
||||||
| NC score | 0.831066 (rank : 36) | θ value | 1.33526e-19 (rank : 39) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP15_MOUSE
|
||||||
| NC score | 0.829130 (rank : 37) | θ value | 5.97985e-28 (rank : 34) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP19_HUMAN
|
||||||
| NC score | 0.827634 (rank : 38) | θ value | 9.56915e-18 (rank : 40) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MMP21_HUMAN
|
||||||
| NC score | 0.784377 (rank : 39) | θ value | 1.63604e-09 (rank : 49) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP21_MOUSE
|
||||||
| NC score | 0.778044 (rank : 40) | θ value | 2.52405e-10 (rank : 47) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.588614 (rank : 41) | θ value | 0.00509761 (rank : 63) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.560333 (rank : 42) | θ value | 0.0252991 (rank : 65) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
HEMO_HUMAN
|
||||||
| NC score | 0.550910 (rank : 43) | θ value | 1.06291 (rank : 102) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02790 | Gene names | HPX | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor (Beta-1B-glycoprotein). | |||||
|
HEMO_MOUSE
|
||||||
| NC score | 0.509711 (rank : 44) | θ value | 0.000786445 (rank : 59) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor. | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.399106 (rank : 45) | θ value | 8.95645e-16 (rank : 42) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.394510 (rank : 46) | θ value | 2.35696e-16 (rank : 41) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.329388 (rank : 47) | θ value | 0.279714 (rank : 84) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MPRI_MOUSE
|
||||||
| NC score | 0.290683 (rank : 48) | θ value | 4.30538e-10 (rank : 48) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
MPRI_HUMAN
|
||||||
| NC score | 0.284525 (rank : 49) | θ value | 2.13673e-09 (rank : 50) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.271796 (rank : 50) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MRC1_MOUSE
|
||||||
| NC score | 0.271068 (rank : 51) | θ value | 4.44505e-15 (rank : 44) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.265533 (rank : 52) | θ value | 2.60593e-15 (rank : 43) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.259987 (rank : 53) | θ value | 1.12514e-34 (rank : 16) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.259449 (rank : 54) | θ value | 1.12514e-34 (rank : 17) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.256119 (rank : 55) | θ value | 3.18553e-13 (rank : 46) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.255808 (rank : 56) | θ value | 1.86753e-13 (rank : 45) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
LY75_HUMAN
|
||||||
| NC score | 0.240768 (rank : 57) | θ value | 4.0297e-08 (rank : 53) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.236931 (rank : 58) | θ value | 1.69304e-06 (rank : 55) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.095197 (rank : 59) | θ value | 3.64472e-09 (rank : 51) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.092726 (rank : 60) | θ value | 4.76016e-09 (rank : 52) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
CP003_HUMAN
|
||||||
| NC score | 0.092672 (rank : 61) | θ value | 0.0330416 (rank : 67) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.091919 (rank : 62) | θ value | 6.87365e-08 (rank : 54) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.082733 (rank : 63) | θ value | 0.000158464 (rank : 58) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
SPRR3_MOUSE
|
||||||
| NC score | 0.076741 (rank : 64) | θ value | 0.0148317 (rank : 64) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O09116 | Gene names | Sprr3 | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta). | |||||
|
CD302_HUMAN
|
||||||
| NC score | 0.070693 (rank : 65) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.069800 (rank : 66) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
ATS13_HUMAN
|
||||||
| NC score | 0.062115 (rank : 67) | θ value | 0.813845 (rank : 95) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q76LX8, Q6UY16, Q710F6, Q711T8, Q96L37, Q9H0G3 | Gene names | ADAMTS13, C9orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CHODL_HUMAN
|
||||||
| NC score | 0.056807 (rank : 68) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| NC score | 0.056647 (rank : 69) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.052661 (rank : 70) | θ value | 0.279714 (rank : 81) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.052035 (rank : 71) | θ value | 0.279714 (rank : 82) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.051512 (rank : 72) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.051358 (rank : 73) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
EMBP_MOUSE
|
||||||
| NC score | 0.050333 (rank : 74) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.046271 (rank : 75) | θ value | 0.0431538 (rank : 69) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.045559 (rank : 76) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.044238 (rank : 77) | θ value | 0.00175202 (rank : 60) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.043708 (rank : 78) | θ value | 2.21117e-06 (rank : 56) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.040901 (rank : 79) | θ value | 0.163984 (rank : 75) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.039356 (rank : 80) | θ value | 7.1131e-05 (rank : 57) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.035834 (rank : 81) | θ value | 0.813845 (rank : 96) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.035728 (rank : 82) | θ value | 0.21417 (rank : 78) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
TSNA1_HUMAN
|
||||||
| NC score | 0.035416 (rank : 83) | θ value | 0.21417 (rank : 79) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NA8 | Gene names | TSNARE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | t-SNARE domain-containing protein 1. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.035294 (rank : 84) | θ value | 0.813845 (rank : 97) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.035010 (rank : 85) | θ value | 0.47712 (rank : 88) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.034093 (rank : 86) | θ value | 0.47712 (rank : 89) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.033193 (rank : 87) | θ value | 1.06291 (rank : 104) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.032402 (rank : 88) | θ value | 0.00298849 (rank : 61) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.031751 (rank : 89) | θ value | 0.47712 (rank : 90) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.029641 (rank : 90) | θ value | 2.36792 (rank : 113) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.029507 (rank : 91) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.029263 (rank : 92) | θ value | 0.0563607 (rank : 70) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.028620 (rank : 93) | θ value | 0.21417 (rank : 76) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.028516 (rank : 94) | θ value | 0.62314 (rank : 91) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.027876 (rank : 95) | θ value | 1.38821 (rank : 107) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.027580 (rank : 96) | θ value | 0.0330416 (rank : 66) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.026675 (rank : 97) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.026600 (rank : 98) | θ value | 0.21417 (rank : 77) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.026463 (rank : 99) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.026083 (rank : 100) | θ value | 0.00390308 (rank : 62) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.025093 (rank : 101) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
K0240_MOUSE
|
||||||
| NC score | 0.023427 (rank : 102) | θ value | 0.279714 (rank : 83) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHH5 | Gene names | Kiaa0240 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0240. | |||||
|
ATS2_HUMAN
|
||||||
| NC score | 0.021779 (rank : 103) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.021558 (rank : 104) | θ value | 1.06291 (rank : 100) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
ATS2_MOUSE
|
||||||
| NC score | 0.021547 (rank : 105) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C9W3 | Gene names | Adamts2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.021540 (rank : 106) | θ value | 0.21417 (rank : 80) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.019963 (rank : 107) | θ value | 0.0961366 (rank : 74) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.019580 (rank : 108) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.019402 (rank : 109) | θ value | 0.0961366 (rank : 73) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
ATS3_HUMAN
|
||||||
| NC score | 0.019324 (rank : 110) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O15072, Q9BXZ8 | Gene names | ADAMTS3, KIAA0366 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-3 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 3) (ADAM-TS 3) (ADAM-TS3) (Procollagen II amino propeptide-processing enzyme) (Procollagen II N-proteinase) (PC II-NP). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.018648 (rank : 111) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.018565 (rank : 112) | θ value | 0.365318 (rank : 85) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.018364 (rank : 113) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.017774 (rank : 114) | θ value | 1.06291 (rank : 101) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.016667 (rank : 115) | θ value | 2.36792 (rank : 112) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.016315 (rank : 116) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
DOS_HUMAN
|
||||||
| NC score | 0.015868 (rank : 117) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.015443 (rank : 118) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
3BHS6_MOUSE
|
||||||
| NC score | 0.015317 (rank : 119) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35469 | Gene names | Hsd3b6 | |||
|
Domain Architecture |

|
|||||
| Description | 3 beta-hydroxysteroid dehydrogenase/delta 5-->4-isomerase type VI (3Beta-HSD VI) [Includes: 3-beta-hydroxy-delta(5)-steroid dehydrogenase (EC 1.1.1.145) (3-beta-hydroxy-5-ene steroid dehydrogenase) (Progesterone reductase); Steroid delta-isomerase (EC 5.3.3.1) (Delta-5-3-ketosteroid isomerase)]. | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.015262 (rank : 120) | θ value | 0.62314 (rank : 92) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
EPOR_HUMAN
|
||||||
| NC score | 0.013821 (rank : 121) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19235, Q15443 | Gene names | EPOR | |||
|
Domain Architecture |

|
|||||
| Description | Erythropoietin receptor precursor (EPO-R). | |||||
|
ATS4_MOUSE
|
||||||
| NC score | 0.013448 (rank : 122) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BNJ2, Q8K384 | Gene names | Adamts4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-4 precursor (EC 3.4.24.82) (A disintegrin and metalloproteinase with thrombospondin motifs 4) (ADAM-TS 4) (ADAM-TS4) (Aggrecanase-1). | |||||
|
ATS4_HUMAN
|
||||||
| NC score | 0.013270 (rank : 123) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75173, Q5VTW2, Q6P4Q8, Q6UWA8, Q9UN83 | Gene names | ADAMTS4, KIAA0688 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-4 precursor (EC 3.4.24.82) (A disintegrin and metalloproteinase with thrombospondin motifs 4) (ADAM-TS 4) (ADAM-TS4) (Aggrecanase-1) (ADMP-1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.013202 (rank : 124) | θ value | 1.38821 (rank : 106) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.012629 (rank : 125) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.012554 (rank : 126) | θ value | 0.62314 (rank : 94) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.012533 (rank : 127) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.012212 (rank : 128) | θ value | 0.0736092 (rank : 72) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.012164 (rank : 129) | θ value | 1.06291 (rank : 103) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.011952 (rank : 130) | θ value | 1.81305 (rank : 110) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.011666 (rank : 131) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.011233 (rank : 132) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.010841 (rank : 133) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.010403 (rank : 134) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.009896 (rank : 135) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.009256 (rank : 136) | θ value | 2.36792 (rank : 114) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.008660 (rank : 137) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
ADA15_MOUSE
|
||||||
| NC score | 0.008452 (rank : 138) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O88839, Q3U7C2, Q8C7Z0, Q91VS9, Q9QYL2 | Gene names | Adam15, Mdc15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin) (AD56). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.008309 (rank : 139) | θ value | 0.813845 (rank : 98) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.008297 (rank : 140) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
FTS1_HUMAN
|
||||||
| NC score | 0.007579 (rank : 141) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8T0 | Gene names | FTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fused toes protein homolog (Ft1). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.007517 (rank : 142) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.006518 (rank : 143) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.006303 (rank : 144) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.004575 (rank : 145) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
VASN_MOUSE
|
||||||
| NC score | 0.004433 (rank : 146) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CZT5, Q8BJJ0, Q8R2G5 | Gene names | Vasn, Slitl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.002650 (rank : 147) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.002545 (rank : 148) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
CRRY_MOUSE
|
||||||
| NC score | 0.002075 (rank : 149) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64735, Q3TV30, Q58E68, Q61447, Q61449, Q8CF59, Q8K328 | Gene names | Crry, Cry | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement regulatory protein Crry precursor (Protein p65). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.000802 (rank : 150) | θ value | 0.47712 (rank : 87) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
KPCD3_HUMAN
|
||||||
| NC score | 0.000313 (rank : 151) | θ value | 0.0330416 (rank : 68) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94806 | Gene names | PRKD3, EPK2, PRKCN | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D3 (EC 2.7.11.13) (Protein kinase C nu type) (nPKC-nu) (Protein kinase EPK2). | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.000157 (rank : 152) | θ value | 0.47712 (rank : 86) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.000085 (rank : 153) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
KPCD3_MOUSE
|
||||||
| NC score | 0.000043 (rank : 154) | θ value | 0.0736092 (rank : 71) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K1Y2 | Gene names | Prkd3, Prkcn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase D3 (EC 2.7.11.13) (Protein kinase C nu type) (nPKC-nu). | |||||
|
GLI1_MOUSE
|
||||||
| NC score | -0.000709 (rank : 155) | θ value | 0.62314 (rank : 93) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.000889 (rank : 156) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | -0.001476 (rank : 157) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ZBT45_HUMAN
|
||||||
| NC score | -0.002287 (rank : 158) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | -0.003911 (rank : 159) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 151 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||