Please be patient as the page loads
|
CP003_HUMAN
|
||||||
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CP003_HUMAN
|
||||||
| θ value | 7.47731e-63 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
SPR2D_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 2) | NC score | 0.143343 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
SPR2I_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 3) | NC score | 0.160559 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 4) | NC score | 0.130462 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 5) | NC score | 0.214262 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.092605 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.158073 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.092672 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.119851 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.174496 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.176166 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
FOXD2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.044067 (rank : 73) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35392 | Gene names | Foxd2, Mf2 | |||
|
Domain Architecture |
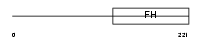
|
|||||
| Description | Forkhead box protein D2 (Mesoderm/mesenchyme forkhead 2) (MF-2). | |||||
|
SPR2F_MOUSE
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.113078 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
SPR2E_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.118459 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.079214 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
VWF_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.118071 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.162463 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.074932 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
SMBT2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.056592 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
SPR2G_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.076961 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
KR122_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.105569 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.039720 (rank : 74) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
FOXD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.037051 (rank : 75) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |
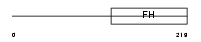
|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
KR106_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.082141 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.120609 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
STK39_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.009550 (rank : 87) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
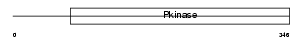
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.049913 (rank : 68) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
KR105_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.092374 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.045027 (rank : 72) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
KR108_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.089054 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
BRSK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.006168 (rank : 88) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.078014 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.076091 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR102_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.088054 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR510_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.051673 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
MT1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.046156 (rank : 71) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13640, P80296 | Gene names | MT1G, MT1K, MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1G (MT-1G) (Metallothionein-IG) (MT-IG) (Metallothionein-1K) (MT-1K). | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.093372 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.060497 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.070490 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.078635 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
SPR2F_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.049460 (rank : 69) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.018655 (rank : 85) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.019471 (rank : 84) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DGKK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.026014 (rank : 78) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.024420 (rank : 81) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.048242 (rank : 70) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRR13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.074341 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
TRBM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.022876 (rank : 82) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |
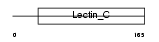
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.010125 (rank : 86) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CY24A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.036182 (rank : 76) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13498, Q14090, Q9BR72 | Gene names | CYBA | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 light chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 22 kDa polypeptide) (p22-phox) (p22phox) (Cytochrome b(558) alpha chain) (Cytochrome b558 subunit alpha) (Superoxide- generating NADPH oxidase light chain subunit). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.028289 (rank : 77) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.022373 (rank : 83) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.025613 (rank : 79) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
AMHR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.025422 (rank : 80) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.063404 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.080191 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052696 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.068723 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.053582 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
GRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.051724 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
GRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.051543 (rank : 66) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.067636 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.089474 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.070553 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.086038 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.078034 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR111_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.064863 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
KR121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.050550 (rank : 67) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
KR123_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.080742 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KR124_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.095420 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
KR261_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.057649 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PEX3 | Gene names | KRTAP26-1, KAP26.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 26-1. | |||||
|
KRA24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.111721 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.061936 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.093173 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.062427 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
PCAF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053094 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.167506 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.156492 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.159602 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.149213 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.150346 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR2J_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.129935 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
SREC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.054580 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
WFDC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051725 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052488 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.066535 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.082590 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ZNFX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.079269 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
CP003_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.47731e-63 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.214262 (rank : 2) | θ value | 0.0252991 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.176166 (rank : 3) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.174496 (rank : 4) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.167506 (rank : 5) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.162463 (rank : 6) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
SPR2I_MOUSE
|
||||||
| NC score | 0.160559 (rank : 7) | θ value | 0.00869519 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70560 | Gene names | Sprr2i | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2I. | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.159602 (rank : 8) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.158073 (rank : 9) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.156492 (rank : 10) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SPR2A_MOUSE
|
||||||
| NC score | 0.150346 (rank : 11) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQK8, O70553 | Gene names | Sprr2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2A. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.149213 (rank : 12) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR2D_MOUSE
|
||||||
| NC score | 0.143343 (rank : 13) | θ value | 0.00390308 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70555 | Gene names | Sprr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2D. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.130462 (rank : 14) | θ value | 0.0113563 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
SPR2J_MOUSE
|
||||||
| NC score | 0.129935 (rank : 15) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70561 | Gene names | Sprr2j | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2J. | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.120609 (rank : 16) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.119851 (rank : 17) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
SPR2E_MOUSE
|
||||||
| NC score | 0.118459 (rank : 18) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70556 | Gene names | Sprr2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2E. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.118071 (rank : 19) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
SPR2F_MOUSE
|
||||||
| NC score | 0.113078 (rank : 20) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70557 | Gene names | Sprr2f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F. | |||||
|
KRA24_HUMAN
|
||||||
| NC score | 0.111721 (rank : 21) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYR9, Q495J2 | Gene names | KRTAP2-4, KAP2.4, KRTAP2.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 2-4 (Keratin-associated protein 2.4) (High sulfur keratin-associated protein 2.4). | |||||
|
KR122_HUMAN
|
||||||
| NC score | 0.105569 (rank : 22) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
KR124_HUMAN
|
||||||
| NC score | 0.095420 (rank : 23) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.093372 (rank : 24) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.093173 (rank : 25) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.092672 (rank : 26) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.092605 (rank : 27) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.092374 (rank : 28) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.089474 (rank : 29) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.089054 (rank : 30) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.088054 (rank : 31) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR10A_HUMAN
|
||||||
| NC score | 0.086038 (rank : 32) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60014 | Gene names | KRTAP10-10, KAP10.10, KAP18-10, KRTAP10.10, KRTAP18-1, KRTAP18.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-10 (Keratin-associated protein 10.10) (High sulfur keratin-associated protein 10.10) (Keratin-associated protein 18-10) (Keratin-associated protein 18.10). | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.082590 (rank : 33) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.082141 (rank : 34) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR123_HUMAN
|
||||||
| NC score | 0.080742 (rank : 35) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P60328 | Gene names | KRTAP12-3, KAP12.3, KRTAP12.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-3 (Keratin-associated protein 12.3) (High sulfur keratin-associated protein 12.3). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.080191 (rank : 36) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
ZNFX1_MOUSE
|
||||||
| NC score | 0.079269 (rank : 37) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R151, Q3U016, Q3UD71, Q3UEZ0 | Gene names | ZNFX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.079214 (rank : 38) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.078635 (rank : 39) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.078034 (rank : 40) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.078014 (rank : 41) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
SPR2G_MOUSE
|
||||||
| NC score | 0.076961 (rank : 42) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70558 | Gene names | Sprr2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2G. | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.076091 (rank : 43) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.074932 (rank : 44) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.074341 (rank : 45) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.070553 (rank : 46) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.070490 (rank : 47) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.068723 (rank : 48) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.067636 (rank : 49) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.066535 (rank : 50) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KR111_HUMAN
|
||||||
| NC score | 0.064863 (rank : 51) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IUC1 | Gene names | KRTAP11-1, KAP11.1, KRTAP11.1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin-associated protein 11-1 (High sulfur keratin-associated protein 11.1). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.063404 (rank : 52) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.062427 (rank : 53) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.061936 (rank : 54) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.060497 (rank : 55) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
KR261_HUMAN
|
||||||
| NC score | 0.057649 (rank : 56) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PEX3 | Gene names | KRTAP26-1, KAP26.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 26-1. | |||||
|
SMBT2_MOUSE
|
||||||
| NC score | 0.056592 (rank : 57) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.054580 (rank : 58) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.053582 (rank : 59) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
PCAF_HUMAN
|
||||||
| NC score | 0.053094 (rank : 60) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.052696 (rank : 61) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.052488 (rank : 62) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
WFDC3_HUMAN
|
||||||
| NC score | 0.051725 (rank : 63) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUB2, Q8TC52, Q9BQP3, Q9BQP4 | Gene names | WFDC3, WAP14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WAP four-disulfide core domain protein 3 precursor (Putative protease inhibitor WAP14). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.051724 (rank : 64) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.051673 (rank : 65) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.051543 (rank : 66) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
KR121_HUMAN
|
||||||
| NC score | 0.050550 (rank : 67) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59990 | Gene names | KRTAP12-1, KAP12.1, KRTAP12.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-1 (Keratin-associated protein 12.1) (High sulfur keratin-associated protein 12.1). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.049913 (rank : 68) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SPR2F_HUMAN
|
||||||
| NC score | 0.049460 (rank : 69) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RM1 | Gene names | SPRR2F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2F (SPR-2F). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.048242 (rank : 70) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MT1G_HUMAN
|
||||||
| NC score | 0.046156 (rank : 71) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13640, P80296 | Gene names | MT1G, MT1K, MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1G (MT-1G) (Metallothionein-IG) (MT-IG) (Metallothionein-1K) (MT-1K). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.045027 (rank : 72) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
FOXD2_MOUSE
|
||||||
| NC score | 0.044067 (rank : 73) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35392 | Gene names | Foxd2, Mf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D2 (Mesoderm/mesenchyme forkhead 2) (MF-2). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.039720 (rank : 74) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
FOXD2_HUMAN
|
||||||
| NC score | 0.037051 (rank : 75) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60548 | Gene names | FOXD2, FKHL17, FREAC9 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D2 (Forkhead-related protein FKHL17) (Forkhead- related transcription factor 9) (FREAC-9). | |||||
|
CY24A_HUMAN
|
||||||
| NC score | 0.036182 (rank : 76) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13498, Q14090, Q9BR72 | Gene names | CYBA | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 light chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 22 kDa polypeptide) (p22-phox) (p22phox) (Cytochrome b(558) alpha chain) (Cytochrome b558 subunit alpha) (Superoxide- generating NADPH oxidase light chain subunit). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.028289 (rank : 77) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
DGKK_HUMAN
|
||||||
| NC score | 0.026014 (rank : 78) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5KSL6 | Gene names | DGKK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase kappa (EC 2.7.1.107) (Diglyceride kinase kappa) (DGK-kappa) (DAG kinase kappa) (142 kDa diacylglycerol kinase). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.025613 (rank : 79) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
AMHR2_HUMAN
|
||||||
| NC score | 0.025422 (rank : 80) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16671, Q13762 | Gene names | AMHR2, AMHR | |||
|
Domain Architecture |

|
|||||
| Description | Anti-Muellerian hormone type-2 receptor precursor (EC 2.7.11.30) (Anti-Muellerian hormone type II receptor) (AMH type II receptor) (MIS type II receptor) (MISRII) (MRII). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.024420 (rank : 81) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
TRBM_HUMAN
|
||||||
| NC score | 0.022876 (rank : 82) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.022373 (rank : 83) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.019471 (rank : 84) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.018655 (rank : 85) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.010125 (rank : 86) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
STK39_MOUSE
|
||||||
| NC score | 0.009550 (rank : 87) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
BRSK1_HUMAN
|
||||||
| NC score | 0.006168 (rank : 88) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDC3, Q5J5B5, Q8NDD0, Q8NDR4, Q8TDC2, Q96AV4, Q96JL4 | Gene names | BRSK1, KIAA1811, SAD1 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 1 (EC 2.7.11.1) (SAD1 kinase) (SAD1A). | |||||