Please be patient as the page loads
|
MT1G_HUMAN
|
||||||
| SwissProt Accessions | P13640, P80296 | Gene names | MT1G, MT1K, MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1G (MT-1G) (Metallothionein-IG) (MT-IG) (Metallothionein-1K) (MT-1K). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MT1M_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 1) | NC score | 0.902339 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N339, Q8TDN3 | Gene names | MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1M (MT-1M) (Metallothionein-IM) (MT-IM). | |||||
|
MT1I_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 2) | NC score | 0.926590 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P80295 | Gene names | MT1I | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1I (MT-1I) (Metallothionein-II). | |||||
|
MT1F_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 3) | NC score | 0.923972 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04733, Q9UI97 | Gene names | MT1F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1F (MT-1F) (Metallothionein-IF) (MT-IF). | |||||
|
MT1E_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 4) | NC score | 0.908109 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04732, Q86YX4 | Gene names | MT1E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1E (MT-1E) (Metallothionein-IE) (MT-IE). | |||||
|
MT2_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 5) | NC score | 0.928633 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P02795, Q14823, Q2HXR9, Q53XT9 | Gene names | MT2A, CES1, MT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-2 (MT-2) (Metallothionein-II) (MT-II) (Metallothionein-2A). | |||||
|
MT1G_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P13640, P80296 | Gene names | MT1G, MT1K, MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1G (MT-1G) (Metallothionein-IG) (MT-IG) (Metallothionein-1K) (MT-1K). | |||||
|
MT1X_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 7) | NC score | 0.894417 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P80297 | Gene names | MT1X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1X (MT-1X) (Metallothionein-IX) (MT-IX). | |||||
|
MT1A_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 8) | NC score | 0.931365 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04731 | Gene names | MT1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1A (MT-1A) (Metallothionein-IA) (MT-IA). | |||||
|
MT1H_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 9) | NC score | 0.921980 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P80294 | Gene names | MT1H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1H (MT-1H) (Metallothionein-IH) (MT-IH) (Metallothionein-0) (MT-0). | |||||
|
MT1L_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 10) | NC score | 0.945397 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q93083 | Gene names | MT1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1L (MT-1L) (Metallothionein-IL) (MT-IL). | |||||
|
MT1B_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 11) | NC score | 0.893275 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07438 | Gene names | MT1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1B (MT-1B) (Metallothionein-IB) (MT-IB). | |||||
|
MT1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 12) | NC score | 0.910463 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P02802, Q64485 | Gene names | Mt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1 (MT-1) (Metallothionein-I) (MT-I). | |||||
|
MT2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 13) | NC score | 0.868933 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02798 | Gene names | Mt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-2 (MT-2) (Metallothionein-II) (MT-II). | |||||
|
MT4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.762383 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47944 | Gene names | MT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-4 (MT-4) (Metallothionein-IV) (MT-IV). | |||||
|
MT4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.740733 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47945 | Gene names | Mt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-4 (MT-4) (Metallothionein-IV) (MT-IV). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.144376 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.122298 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
CT127_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.778956 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQN2 | Gene names | C20orf127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative metallothionein C20orf127. | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.118399 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.113870 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.056851 (rank : 141) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.133817 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.117417 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.120246 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
ADA28_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.075534 (rank : 103) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UKQ2, Q9Y339, Q9Y3S0 | Gene names | ADAM28, MDCL | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein-L) (MDC-L) (eMDC II) (ADAM23). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.135530 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.145642 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.054227 (rank : 154) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.125782 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.075611 (rank : 102) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.101965 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
KR124_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.028911 (rank : 184) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.122566 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
SIVA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.125019 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15304, Q96P98, Q9UPD6 | Gene names | SIVA | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
ADA28_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.078150 (rank : 93) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JLN6, Q8K5D2, Q8K5D3 | Gene names | Adam28 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Thymic epithelial cell-ADAM) (TECADAM). | |||||
|
ADAM9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.071555 (rank : 111) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61072, Q60618, Q61853, Q80U94 | Gene names | Adam9, Kiaa0021, Mdc9, Mltng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma). | |||||
|
CP003_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.046156 (rank : 179) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
ITB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.120149 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.139386 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.123345 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.099898 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
ADAM9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.066477 (rank : 118) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13443, Q10718, Q8NFM6 | Gene names | ADAM9, KIAA0021, MCMP, MDC9, MLTNG | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma) (Cellular disintegrin-related protein). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.086711 (rank : 79) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.100467 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
USH2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.115252 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
ADA24_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.049190 (rank : 178) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R160 | Gene names | Adam24 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 24 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 24) (Testase 1). | |||||
|
ITB6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.113926 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.121084 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
TNR9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.123326 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
ADM1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.051401 (rank : 169) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R534, Q9R156 | Gene names | Adam1b | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1b precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1b) (Fertilin alpha 1b subunit). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.015530 (rank : 186) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.006365 (rank : 187) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ITB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.133859 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.110733 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ADA33_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.044758 (rank : 180) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q923W9 | Gene names | Adam33 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 33 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 33). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.059186 (rank : 133) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
ITB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.118675 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.129872 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.104007 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
SEPP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.030620 (rank : 183) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49908, Q6PD59, Q6PI43, Q6PI87, Q6PJF9 | Gene names | SEPP1, SELP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP). | |||||
|
TR10A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.027492 (rank : 185) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
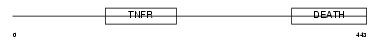
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
TRI42_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.032848 (rank : 181) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
TRI42_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.031645 (rank : 182) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
ADA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.061016 (rank : 127) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 32 | |
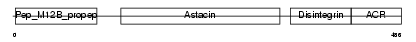
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |
|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.060708 (rank : 128) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.077876 (rank : 94) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0K1, O75075, O75076, Q9P0K2, Q9UIA1, Q9UKK2 | Gene names | ADAM22, MDC2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 22 precursor (A disintegrin and metalloproteinase domain 22) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein 2) (Metalloproteinase-disintegrin ADAM22-3). | |||||
|
ADA22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.083580 (rank : 84) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R1V6, Q5TLI8, Q5TLI9, Q5TLJ0, Q5TLJ1, Q5TLJ2, Q5TLJ3, Q5TLJ4, Q5TLJ5, Q5TLJ6, Q5TLJ7, Q5TLJ8, Q5TLJ9, Q5TLK0, Q5TLK1, Q5TLK2, Q5TLK3, Q5TLK4, Q5TLK5, Q5TLK6, Q5TLK7, Q5TLK8, Q8BSF2, Q9R1V5 | Gene names | Adam22 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 22 precursor (A disintegrin and metalloproteinase domain 22). | |||||
|
ADAM8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.050121 (rank : 176) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.071700 (rank : 110) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.075776 (rank : 101) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.077324 (rank : 97) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
BMPER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.052534 (rank : 162) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054176 (rank : 155) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.051031 (rank : 172) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054336 (rank : 153) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056316 (rank : 143) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.054493 (rank : 152) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.055334 (rank : 146) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.051940 (rank : 165) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 31 | |
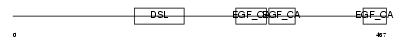
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.055382 (rank : 145) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DNER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.051102 (rank : 171) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
ESM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.051484 (rank : 168) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.086150 (rank : 80) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.085692 (rank : 82) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
ITB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.092599 (rank : 73) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ITB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.095987 (rank : 70) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ITB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.118737 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 43 | |
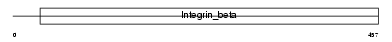
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.065601 (rank : 119) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
ITB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.102366 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.106430 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P26010 | Gene names | ITGB7 | |||
|
Domain Architecture |
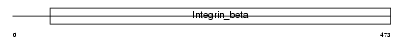
|
|||||
| Description | Integrin beta-7 precursor. | |||||
|
ITB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.108430 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P26011, Q3U2M1, Q64656 | Gene names | Itgb7 | |||
|
Domain Architecture |
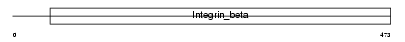
|
|||||
| Description | Integrin beta-7 precursor (Integrin beta-P) (M290 IEL antigen). | |||||
|
ITB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.106350 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.055018 (rank : 149) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054780 (rank : 151) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056103 (rank : 144) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.058687 (rank : 135) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.068672 (rank : 115) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.053486 (rank : 157) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.051990 (rank : 164) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.075359 (rank : 104) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.057012 (rank : 140) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.069899 (rank : 114) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.072418 (rank : 109) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR108_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.058386 (rank : 138) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.060236 (rank : 129) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.059674 (rank : 131) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053526 (rank : 156) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.081930 (rank : 86) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR412_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050647 (rank : 173) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.128336 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.111332 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.051835 (rank : 166) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
KRA13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053092 (rank : 159) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
KRA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052157 (rank : 163) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KRA45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.063116 (rank : 122) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
KRA47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.058597 (rank : 136) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.123775 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.117441 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.117676 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.120764 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.121574 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.115280 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.112754 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.111770 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.129002 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.053185 (rank : 158) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055206 (rank : 147) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.103344 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.088773 (rank : 77) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.097318 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.085690 (rank : 83) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.088464 (rank : 78) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.088818 (rank : 75) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.093489 (rank : 72) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.089249 (rank : 74) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
LAMB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.088775 (rank : 76) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.102543 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.105527 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.105195 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LAMC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.101474 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.055194 (rank : 148) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054819 (rank : 150) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.062823 (rank : 124) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
LCE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.063880 (rank : 121) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.076276 (rank : 98) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.073190 (rank : 108) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.100773 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.096278 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.519749 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
MT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.502829 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P28184 | Gene names | Mt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF). | |||||
|
NAGPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.078683 (rank : 92) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NAGPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.077747 (rank : 95) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050040 (rank : 177) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NET1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.075891 (rank : 100) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95631 | Gene names | NTN1, NTN1L | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
NET1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.075938 (rank : 99) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09118, Q60832, Q9QY50 | Gene names | Ntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
NET2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.079870 (rank : 89) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
NET2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.079473 (rank : 91) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1A3, Q9QY49, Q9WVA6 | Gene names | Ntn2l, Ntn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor (Netrin-3). | |||||
|
NET4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.080438 (rank : 88) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
NET4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.080678 (rank : 87) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JI33 | Gene names | Ntn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.051623 (rank : 167) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.052832 (rank : 160) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.050270 (rank : 174) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050247 (rank : 175) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NTNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.070892 (rank : 113) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.071198 (rank : 112) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.077738 (rank : 96) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96CW9, Q6UXY0, Q96JH0 | Gene names | NTNG2, KIAA1857, LMNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
NTNG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.074681 (rank : 106) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4F1, Q8R4F2, Q8VIP6, Q8VIP7, Q8VIP8 | Gene names | Ntng2, Lmnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
PCSK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.073211 (rank : 107) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.067752 (rank : 116) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.067180 (rank : 117) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SIVA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.051264 (rank : 170) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54926, Q3U6J2, Q921I8, Q9CWL1, Q9R1Q0, Q9R1Q1 | Gene names | Siva | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.058573 (rank : 137) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.056759 (rank : 142) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.059934 (rank : 130) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.059095 (rank : 134) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.061226 (rank : 125) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TENA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.057990 (rank : 139) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.059669 (rank : 132) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TENX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.075205 (rank : 105) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
TNR7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.079681 (rank : 90) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |
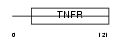
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
TNR7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.083508 (rank : 85) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
TNR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.085768 (rank : 81) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |
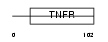
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
USH2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.095389 (rank : 71) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052774 (rank : 161) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.061190 (rank : 126) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.063068 (rank : 123) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.064266 (rank : 120) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
MT1G_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.26297e-08 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P13640, P80296 | Gene names | MT1G, MT1K, MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1G (MT-1G) (Metallothionein-IG) (MT-IG) (Metallothionein-1K) (MT-1K). | |||||
|
MT1L_HUMAN
|
||||||
| NC score | 0.945397 (rank : 2) | θ value | 4.1701e-05 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q93083 | Gene names | MT1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1L (MT-1L) (Metallothionein-IL) (MT-IL). | |||||
|
MT1A_HUMAN
|
||||||
| NC score | 0.931365 (rank : 3) | θ value | 3.19293e-05 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04731 | Gene names | MT1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1A (MT-1A) (Metallothionein-IA) (MT-IA). | |||||
|
MT2_HUMAN
|
||||||
| NC score | 0.928633 (rank : 4) | θ value | 2.79066e-09 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P02795, Q14823, Q2HXR9, Q53XT9 | Gene names | MT2A, CES1, MT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-2 (MT-2) (Metallothionein-II) (MT-II) (Metallothionein-2A). | |||||
|
MT1I_HUMAN
|
||||||
| NC score | 0.926590 (rank : 5) | θ value | 1.9326e-10 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P80295 | Gene names | MT1I | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1I (MT-1I) (Metallothionein-II). | |||||
|
MT1F_HUMAN
|
||||||
| NC score | 0.923972 (rank : 6) | θ value | 7.34386e-10 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04733, Q9UI97 | Gene names | MT1F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1F (MT-1F) (Metallothionein-IF) (MT-IF). | |||||
|
MT1H_HUMAN
|
||||||
| NC score | 0.921980 (rank : 7) | θ value | 4.1701e-05 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P80294 | Gene names | MT1H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1H (MT-1H) (Metallothionein-IH) (MT-IH) (Metallothionein-0) (MT-0). | |||||
|
MT1_MOUSE
|
||||||
| NC score | 0.910463 (rank : 8) | θ value | 0.00102713 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P02802, Q64485 | Gene names | Mt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1 (MT-1) (Metallothionein-I) (MT-I). | |||||
|
MT1E_HUMAN
|
||||||
| NC score | 0.908109 (rank : 9) | θ value | 2.13673e-09 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P04732, Q86YX4 | Gene names | MT1E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1E (MT-1E) (Metallothionein-IE) (MT-IE). | |||||
|
MT1M_HUMAN
|
||||||
| NC score | 0.902339 (rank : 10) | θ value | 1.47974e-10 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N339, Q8TDN3 | Gene names | MT1M | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1M (MT-1M) (Metallothionein-IM) (MT-IM). | |||||
|
MT1X_HUMAN
|
||||||
| NC score | 0.894417 (rank : 11) | θ value | 1.17247e-07 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P80297 | Gene names | MT1X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1X (MT-1X) (Metallothionein-IX) (MT-IX). | |||||
|
MT1B_HUMAN
|
||||||
| NC score | 0.893275 (rank : 12) | θ value | 0.00102713 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07438 | Gene names | MT1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-1B (MT-1B) (Metallothionein-IB) (MT-IB). | |||||
|
MT2_MOUSE
|
||||||
| NC score | 0.868933 (rank : 13) | θ value | 0.00175202 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P02798 | Gene names | Mt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-2 (MT-2) (Metallothionein-II) (MT-II). | |||||
|
CT127_HUMAN
|
||||||
| NC score | 0.778956 (rank : 14) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQN2 | Gene names | C20orf127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative metallothionein C20orf127. | |||||
|
MT4_HUMAN
|
||||||
| NC score | 0.762383 (rank : 15) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47944 | Gene names | MT4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-4 (MT-4) (Metallothionein-IV) (MT-IV). | |||||
|
MT4_MOUSE
|
||||||
| NC score | 0.740733 (rank : 16) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47945 | Gene names | Mt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-4 (MT-4) (Metallothionein-IV) (MT-IV). | |||||
|
MT3_HUMAN
|
||||||
| NC score | 0.519749 (rank : 17) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
MT3_MOUSE
|
||||||
| NC score | 0.502829 (rank : 18) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P28184 | Gene names | Mt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.145642 (rank : 19) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.144376 (rank : 20) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
ITB5_MOUSE
|
||||||
| NC score | 0.139386 (rank : 21) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.135530 (rank : 22) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.133859 (rank : 23) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.133817 (rank : 24) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
ITB5_HUMAN
|
||||||
| NC score | 0.129872 (rank : 25) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.129002 (rank : 26) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.128336 (rank : 27) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.125782 (rank : 28) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
SIVA_HUMAN
|
||||||
| NC score | 0.125019 (rank : 29) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15304, Q96P98, Q9UPD6 | Gene names | SIVA | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.123775 (rank : 30) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.123345 (rank : 31) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
TNR9_HUMAN
|
||||||
| NC score | 0.123326 (rank : 32) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.122566 (rank : 33) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.122298 (rank : 34) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.121574 (rank : 35) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.121084 (rank : 36) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.120764 (rank : 37) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.120246 (rank : 38) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
ITB1_MOUSE
|
||||||
| NC score | 0.120149 (rank : 39) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB3_MOUSE
|
||||||
| NC score | 0.118737 (rank : 40) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB3_HUMAN
|
||||||
| NC score | 0.118675 (rank : 41) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.118399 (rank : 42) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.117676 (rank : 43) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.117441 (rank : 44) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.117417 (rank : 45) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.115280 (rank : 46) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.115252 (rank : 47) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
ITB6_HUMAN
|
||||||
| NC score | 0.113926 (rank : 48) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.113870 (rank : 49) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.112754 (rank : 50) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.111770 (rank : 51) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.111332 (rank : 52) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.110733 (rank : 53) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ITB7_MOUSE
|
||||||
| NC score | 0.108430 (rank : 54) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P26011, Q3U2M1, Q64656 | Gene names | Itgb7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-7 precursor (Integrin beta-P) (M290 IEL antigen). | |||||
|
ITB7_HUMAN
|
||||||
| NC score | 0.106430 (rank : 55) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P26010 | Gene names | ITGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-7 precursor. | |||||
|
ITB8_HUMAN
|
||||||
| NC score | 0.106350 (rank : 56) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
LAMC1_MOUSE
|
||||||
| NC score | 0.105527 (rank : 57) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.105195 (rank : 58) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.104007 (rank : 59) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.103344 (rank : 60) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.102543 (rank : 61) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.102366 (rank : 62) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.101965 (rank : 63) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
LAMC3_MOUSE
|
||||||
| NC score | 0.101474 (rank : 64) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.100773 (rank : 65) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.100467 (rank : 66) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.099898 (rank : 67) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
LAMA4_MOUSE
|
||||||
| NC score | 0.097318 (rank : 68) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.096278 (rank : 69) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.095987 (rank : 70) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.095389 (rank : 71) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.093489 (rank : 72) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
ITB2_HUMAN
|
||||||
| NC score | 0.092599 (rank : 73) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.089249 (rank : 74) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
LAMB2_HUMAN
|
||||||
| NC score | 0.088818 (rank : 75) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
LAMB3_MOUSE
|
||||||
| NC score | 0.088775 (rank : 76) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.088773 (rank : 77) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.088464 (rank : 78) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.086711 (rank : 79) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.086150 (rank : 80) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
TNR9_MOUSE
|
||||||
| NC score | 0.085768 (rank : 81) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.085692 (rank : 82) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.085690 (rank : 83) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
ADA22_MOUSE
|
||||||
| NC score | 0.083580 (rank : 84) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R1V6, Q5TLI8, Q5TLI9, Q5TLJ0, Q5TLJ1, Q5TLJ2, Q5TLJ3, Q5TLJ4, Q5TLJ5, Q5TLJ6, Q5TLJ7, Q5TLJ8, Q5TLJ9, Q5TLK0, Q5TLK1, Q5TLK2, Q5TLK3, Q5TLK4, Q5TLK5, Q5TLK6, Q5TLK7, Q5TLK8, Q8BSF2, Q9R1V5 | Gene names | Adam22 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 22 precursor (A disintegrin and metalloproteinase domain 22). | |||||
|
TNR7_MOUSE
|
||||||
| NC score | 0.083508 (rank : 85) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.081930 (rank : 86) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
NET4_MOUSE
|
||||||
| NC score | 0.080678 (rank : 87) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JI33 | Gene names | Ntn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin). | |||||
|
NET4_HUMAN
|
||||||
| NC score | 0.080438 (rank : 88) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
NET2_HUMAN
|
||||||
| NC score | 0.079870 (rank : 89) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
TNR7_HUMAN
|
||||||
| NC score | 0.079681 (rank : 90) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
NET2_MOUSE
|
||||||
| NC score | 0.079473 (rank : 91) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9R1A3, Q9QY49, Q9WVA6 | Gene names | Ntn2l, Ntn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor (Netrin-3). | |||||
|
NAGPA_HUMAN
|
||||||
| NC score | 0.078683 (rank : 92) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UK23, Q96EJ8 | Gene names | NAGPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
ADA28_MOUSE
|
||||||
| NC score | 0.078150 (rank : 93) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JLN6, Q8K5D2, Q8K5D3 | Gene names | Adam28 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Thymic epithelial cell-ADAM) (TECADAM). | |||||
|
ADA22_HUMAN
|
||||||
| NC score | 0.077876 (rank : 94) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P0K1, O75075, O75076, Q9P0K2, Q9UIA1, Q9UKK2 | Gene names | ADAM22, MDC2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 22 precursor (A disintegrin and metalloproteinase domain 22) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein 2) (Metalloproteinase-disintegrin ADAM22-3). | |||||
|
NAGPA_MOUSE
|
||||||
| NC score | 0.077747 (rank : 95) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NTNG2_HUMAN
|
||||||
| NC score | 0.077738 (rank : 96) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96CW9, Q6UXY0, Q96JH0 | Gene names | NTNG2, KIAA1857, LMNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.077324 (rank : 97) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.076276 (rank : 98) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NET1_MOUSE
|
||||||
| NC score | 0.075938 (rank : 99) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O09118, Q60832, Q9QY50 | Gene names | Ntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
NET1_HUMAN
|
||||||
| NC score | 0.075891 (rank : 100) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95631 | Gene names | NTN1, NTN1L | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.075776 (rank : 101) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.075611 (rank : 102) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ADA28_HUMAN
|
||||||
| NC score | 0.075534 (rank : 103) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UKQ2, Q9Y339, Q9Y3S0 | Gene names | ADAM28, MDCL | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein-L) (MDC-L) (eMDC II) (ADAM23). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.075359 (rank : 104) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.075205 (rank : 105) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
NTNG2_MOUSE
|
||||||
| NC score | 0.074681 (rank : 106) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4F1, Q8R4F2, Q8VIP6, Q8VIP7, Q8VIP8 | Gene names | Ntng2, Lmnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
PCSK6_HUMAN
|
||||||
| NC score | 0.073211 (rank : 107) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.073190 (rank : 108) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.072418 (rank : 109) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.071700 (rank : 110) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ADAM9_MOUSE
|
||||||
| NC score | 0.071555 (rank : 111) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61072, Q60618, Q61853, Q80U94 | Gene names | Adam9, Kiaa0021, Mdc9, Mltng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma). | |||||
|
NTNG1_MOUSE
|
||||||
| NC score | 0.071198 (rank : 112) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_HUMAN
|
||||||
| NC score | 0.070892 (rank : 113) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.069899 (rank : 114) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.068672 (rank : 115) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.067752 (rank : 116) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.067180 (rank : 117) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ADAM9_HUMAN
|
||||||
| NC score | 0.066477 (rank : 118) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13443, Q10718, Q8NFM6 | Gene names | ADAM9, KIAA0021, MCMP, MDC9, MLTNG | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma) (Cellular disintegrin-related protein). | |||||
|
ITB4_HUMAN
|
||||||
| NC score | 0.065601 (rank : 119) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P16144, O14690, O14691, O15339, O15340, O15341, Q9UIQ4 | Gene names | ITGB4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-4 precursor (GP150) (CD104 antigen). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.064266 (rank : 120) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
LCE3A_HUMAN
|
||||||
| NC score | 0.063880 (rank : 121) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA76 | Gene names | LCE3A, LEP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 3A (Late envelope protein 13). | |||||
|
KRA45_HUMAN
|
||||||
| NC score | 0.063116 (rank : 122) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYR2 | Gene names | KRTAP4-5, KAP4.5, KRTAP4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh sulfur keratin-associated protein 4.5). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.063068 (rank : 123) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.062823 (rank : 124) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.061226 (rank : 125) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.061190 (rank : 126) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
ADA11_HUMAN
|
||||||
| NC score | 0.061016 (rank : 127) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| NC score | 0.060708 (rank : 128) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.060236 (rank : 129) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.059934 (rank : 130) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.059674 (rank : 131) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.059669 (rank : 132) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.059186 (rank : 133) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.059095 (rank : 134) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.058687 (rank : 135) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
KRA47_HUMAN
|
||||||
| NC score | 0.058597 (rank : 136) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BYR0 | Gene names | KRTAP4-7, KAP4.7, KRTAP4.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-7 (Keratin-associated protein 4.7) (Ultrahigh sulfur keratin-associated protein 4.7). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.058573 (rank : 137) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.058386 (rank : 138) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.057990 (rank : 139) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.057012 (rank : 140) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.056851 (rank : 141) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.056759 (rank : 142) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.056316 (rank : 143) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.056103 (rank : 144) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.055382 (rank : 145) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
DLL3_MOUSE
|
||||||
| NC score | 0.055334 (rank : 146) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88516, O35675, Q80W06, Q9QWL9, Q9QWZ7 | Gene names | Dll3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3) (M-Delta- 3). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.055206 (rank : 147) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.055194 (rank : 148) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.055018 (rank : 149) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.054819 (rank : 150) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.054780 (rank : 151) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.054493 (rank : 152) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.054336 (rank : 153) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.054227 (rank : 154) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.054176 (rank : 155) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.053526 (rank : 156) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.053486 (rank : 157) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.053185 (rank : 158) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA13_HUMAN
|
||||||
| NC score | 0.053092 (rank : 159) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUG1, Q07628, Q8IUG0, Q9BYS2 | Gene names | KRTAP1-3, B2B, KAP1.2, KAP1.3, KAP1.8, KAP1.9, KRATP1.9, KRTAP1.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-3 (Keratin-associated protein 1.8) (Keratin-associated protein 1.9). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.052832 (rank : 160) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.052774 (rank : 161) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.052534 (rank : 162) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
KRA15_HUMAN
|
||||||
| NC score | 0.052157 (rank : 163) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BYS1, Q52LP6 | Gene names | KRTAP1-5, KAP1.5, KRTAP1.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-5 (Keratin-associated protein 1.5) (High sulfur keratin-associated protein 1.5). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.051990 (rank : 164) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.051940 (rank : 165) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.051835 (rank : 166) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.051623 (rank : 167) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.051484 (rank : 168) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
ADM1B_MOUSE
|
||||||
| NC score | 0.051401 (rank : 169) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R534, Q9R156 | Gene names | Adam1b | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1b precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1b) (Fertilin alpha 1b subunit). | |||||
|
SIVA_MOUSE
|
||||||
| NC score | 0.051264 (rank : 170) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54926, Q3U6J2, Q921I8, Q9CWL1, Q9R1Q0, Q9R1Q1 | Gene names | Siva | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
DNER_HUMAN
|
||||||
| NC score | 0.051102 (rank : 171) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NFT8, Q53R88, Q53TP7, Q53TQ5, Q8IYT0, Q8TB42, Q9NTF1, Q9UDM2 | Gene names | DNER, BET | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta and Notch-like epidermal growth factor-related receptor precursor. | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.051031 (rank : 172) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
KR412_HUMAN
|
||||||
| NC score | 0.050647 (rank : 173) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BQ66, Q495I0 | Gene names | KRTAP4-12, KAP4.12, KRTAP4.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-12 (Keratin-associated protein 4.12) (Ultrahigh sulfur keratin-associated protein 4.12). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.050270 (rank : 174) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.050247 (rank : 175) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ADAM8_MOUSE
|
||||||
| NC score | 0.050121 (rank : 176) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05910 | Gene names | Adam8, Ms2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 8) (Cell surface antigen MS2) (Macrophage cysteine-rich glycoprotein) (CD156 antigen). | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.050040 (rank : 177) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
ADA24_MOUSE
|
||||||
| NC score | 0.049190 (rank : 178) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9R160 | Gene names | Adam24 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 24 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 24) (Testase 1). | |||||
|
CP003_HUMAN
|
||||||
| NC score | 0.046156 (rank : 179) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95177, O95176 | Gene names | C16orf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf3. | |||||
|
ADA33_MOUSE
|
||||||
| NC score | 0.044758 (rank : 180) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q923W9 | Gene names | Adam33 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 33 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 33). | |||||
|
TRI42_HUMAN
|
||||||
| NC score | 0.032848 (rank : 181) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
TRI42_MOUSE
|
||||||
| NC score | 0.031645 (rank : 182) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
SEPP1_HUMAN
|
||||||
| NC score | 0.030620 (rank : 183) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49908, Q6PD59, Q6PI43, Q6PI87, Q6PJF9 | Gene names | SEPP1, SELP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP). | |||||
|
KR124_HUMAN
|
||||||
| NC score | 0.028911 (rank : 184) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P60329 | Gene names | KRTAP12-4, KAP12.4, KRTAP12.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-4 (Keratin-associated protein 12.4) (High sulfur keratin-associated protein 12.4). | |||||
|
TR10A_HUMAN
|
||||||
| NC score | 0.027492 (rank : 185) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.015530 (rank : 186) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.006365 (rank : 187) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||