Please be patient as the page loads
|
SEL1L_MOUSE
|
||||||
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SEL1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994744 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MMP9_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 3) | NC score | 0.394510 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MMP9_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 4) | NC score | 0.390595 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
MMP2_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 5) | NC score | 0.354355 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 6) | NC score | 0.353886 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
MRC2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 7) | NC score | 0.401992 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 8) | NC score | 0.403567 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 9) | NC score | 0.296858 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 10) | NC score | 0.296624 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 11) | NC score | 0.393023 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 12) | NC score | 0.402620 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
K0141_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 13) | NC score | 0.366345 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCV6, Q6A0B8 | Gene names | Kiaa0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
K0141_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 14) | NC score | 0.357557 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14154, Q969R4, Q96EU9 | Gene names | KIAA0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
MPRI_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 15) | NC score | 0.442330 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
FA12_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 16) | NC score | 0.148875 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
MPRI_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 17) | NC score | 0.433332 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
LY75_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 18) | NC score | 0.384626 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
HGFA_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 19) | NC score | 0.140975 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
LY75_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 20) | NC score | 0.378274 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 21) | NC score | 0.137589 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
EF2K_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 22) | NC score | 0.186391 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 23) | NC score | 0.176522 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |
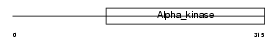
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
WFS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.064115 (rank : 103) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76024, Q8N6I3, Q9UNW6 | Gene names | WFS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.030331 (rank : 150) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
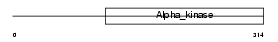
NSD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.022767 (rank : 155) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.027054 (rank : 152) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BMS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.040882 (rank : 149) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
DLG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.017900 (rank : 163) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.023760 (rank : 154) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.010481 (rank : 175) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.024054 (rank : 153) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.020076 (rank : 160) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
DLG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.018218 (rank : 161) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.014876 (rank : 167) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
MARK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | -0.000606 (rank : 184) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.013683 (rank : 170) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.013584 (rank : 171) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
CAYP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.029759 (rank : 151) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.067179 (rank : 93) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.021254 (rank : 158) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
KCC4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | -0.000653 (rank : 185) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08414, Q61381 | Gene names | Camk4 | |||
|
Domain Architecture |
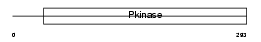
|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.016139 (rank : 166) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
TRUB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.020508 (rank : 159) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95900 | Gene names | TRUB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tRNA pseudouridine synthase 2 (EC 5.4.99.-). | |||||
|
CHIC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.016363 (rank : 165) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VXU3 | Gene names | CHIC1, BRX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
ABCB6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.004733 (rank : 181) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP58, O75542 | Gene names | ABCB6, MTABC3 | |||
|
Domain Architecture |
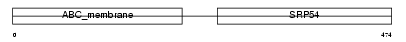
|
|||||
| Description | ATP-binding cassette sub-family B member 6, mitochondrial precursor (Mitochondrial ABC transporter 3) (Mt-ABC transporter 3) (ABC transporter umat). | |||||
|
AN32A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.010999 (rank : 173) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.010999 (rank : 174) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.012271 (rank : 172) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CCNF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.022367 (rank : 157) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.022547 (rank : 156) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
DDX24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.004822 (rank : 180) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.000543 (rank : 183) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.005929 (rank : 179) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.016800 (rank : 164) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.017985 (rank : 162) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
DLG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.013874 (rank : 169) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
DLG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.013992 (rank : 168) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
HEPH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.010459 (rank : 176) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.006593 (rank : 178) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.008297 (rank : 177) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.002209 (rank : 182) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ASGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.053695 (rank : 138) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |
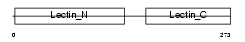
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
ASGR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.058655 (rank : 124) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |
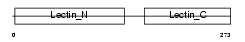
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
ASGR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.060360 (rank : 122) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |
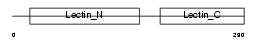
|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
C209A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.062179 (rank : 112) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |
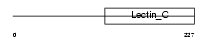
|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
C209B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.062623 (rank : 110) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CJ91, Q8BGZ0, Q8BHK7, Q8CJ86, Q8CJ87, Q8CJ88, Q8CJ89, Q8CJ90, Q8CJ92, Q8CJ93, Q8CJ94, Q91ZW4, Q91ZX0, Q9D8V4 | Gene names | Cd209b | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein B (DC-SIGN-related protein 1) (DC-SIGNR1) (OtB7). | |||||
|
C209C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.064225 (rank : 102) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
C209D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.067519 (rank : 88) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZW8, Q8VIK4 | Gene names | Cd209d | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein D (DC-SIGN-related protein 3) (DC-SIGNR3). | |||||
|
C209E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.063484 (rank : 106) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |
|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
CD209_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.062862 (rank : 109) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
CD302_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.125616 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.125058 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CD69_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.060933 (rank : 117) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |
|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
CD69_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.056115 (rank : 129) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P37217 | Gene names | Cd69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early activation antigen CD69. | |||||
|
CHODL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.101530 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.100867 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |
|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CLC10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.057355 (rank : 128) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
CLC11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.070694 (rank : 72) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y240 | Gene names | CLEC11A, CLECSF3, LSLCL, SCGF | |||
|
Domain Architecture |
|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin) (p47) (C-type lectin superfamily member 3). | |||||
|
CLC11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.063789 (rank : 105) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88200, Q8C946, Q9CTF0 | Gene names | Clec11a, Scgf | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin). | |||||
|
CLC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.054558 (rank : 133) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC01, Q8IUW7, Q9NZH3 | Gene names | CLEC1A, CLEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A (C-type lectin-like receptor 1) (CLEC-1). | |||||
|
CLC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.060559 (rank : 121) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BWY2, Q7TPX7, Q8BMH5 | Gene names | Clec1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A. | |||||
|
CLC3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.065046 (rank : 96) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75596, Q6UXF5 | Gene names | CLEC3A, CLECSF1 | |||
|
Domain Architecture |
|
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.067204 (rank : 92) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPW4 | Gene names | Clec3a, Clecsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.066683 (rank : 94) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UMR7, Q8WXW9, Q9H2Z9, Q9NS33, Q9UI34 | Gene names | CLEC4A, CLECSF6, DCIR, LLIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor) (Lectin-like immunoreceptor) (C-type lectin DDB27). | |||||
|
CLC4A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.067926 (rank : 83) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
CLC4C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.064884 (rank : 98) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
CLC4D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.063046 (rank : 108) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
CLC4D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.068523 (rank : 80) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
CLC4E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.068592 (rank : 79) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.067373 (rank : 90) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.072296 (rank : 67) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.076279 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
CLC4K_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.064966 (rank : 97) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4K_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.060898 (rank : 120) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4M_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.060986 (rank : 115) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
CLC6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.067718 (rank : 85) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CLC6A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.066535 (rank : 95) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKF4, Q9JKF2, Q9JKF3 | Gene names | Clec6a, Clecsf10, Dectin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CLC9A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.060926 (rank : 118) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UXN8 | Gene names | CLEC9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
CLC9A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.053965 (rank : 137) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRU4 | Gene names | Clec9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.064640 (rank : 99) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.069138 (rank : 78) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.067538 (rank : 86) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
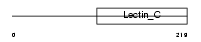
EMBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.081212 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13727, Q14227 | Gene names | PRG2, MBP | |||
|
Domain Architecture |
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (EMBP) (Pregnancy-associated major basic protein) (Proteoglycan 2, bone marrow) (BMPG). | |||||
|
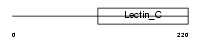
EMBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.087991 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
FCER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.079238 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
FCER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.076108 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
FREM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.050559 (rank : 147) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q684R7, Q5H8C2, Q5M7B3, Q8C732 | Gene names | Frem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
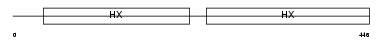
HEMO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.054391 (rank : 135) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |
|
|||||
| Description | Hemopexin precursor. | |||||
|
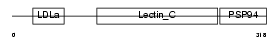
IDD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051906 (rank : 143) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
KLRBA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.060353 (rank : 123) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27811 | Gene names | Klrb1a, Ly55, Ly55a | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell surface protein P1-2 (NKR-P1 2) (NKR-P1.7). | |||||
|
KLRBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.061963 (rank : 113) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27812 | Gene names | Klrb1b, Ly55-b, Ly55b | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell surface protein P1-34 (NKR-P1 34). | |||||
|
KLRBC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.058177 (rank : 127) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27814 | Gene names | Klrb1c, Ly55-c, Ly55c | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cell surface protein P1-40 (NKR-P1 40) (NKR-P1.9). | |||||
|
KLRD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.070099 (rank : 74) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13241, O43321, O43773, Q9UBE3, Q9UEQ0 | Gene names | KLRD1, CD94 | |||
|
Domain Architecture |
|
|||||
| Description | Natural killer cells antigen CD94 (NK cell receptor) (Killer cell lectin-like receptor subfamily D member 1) (KP43). | |||||
|
KLRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.075070 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZS2, Q96PR2, Q96PR3, Q9NZS1 | Gene names | KLRF1, ML | |||
|
Domain Architecture |
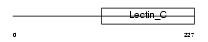
|
|||||
| Description | Killer cell lectin-like receptor subfamily F member 1 (Lectin-like receptor F1) (Activating coreceptor NKp80). | |||||
|
LIT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.071674 (rank : 68) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43137 | Gene names | Reg1 | |||
|
Domain Architecture |
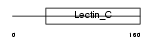
|
|||||
| Description | Lithostathine 1 precursor (Pancreatic stone protein 1) (PSP) (Pancreatic thread protein 1) (PTP) (Islet of Langerhans regenerating protein 1) (REG 1). | |||||
|
LIT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.076651 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08731 | Gene names | Reg2 | |||
|
Domain Architecture |
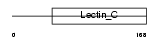
|
|||||
| Description | Lithostathine 2 precursor (Pancreatic stone protein 2) (PSP) (Pancreatic thread protein 2) (PTP) (Islet of Langerhans regenerating protein 2) (REG 2). | |||||
|
LYAM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.054406 (rank : 134) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |
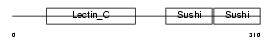
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
LYAM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.054256 (rank : 136) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |
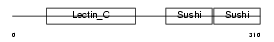
|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
MBL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.052548 (rank : 140) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39039 | Gene names | Mbl1 | |||
|
Domain Architecture |
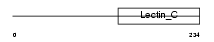
|
|||||
| Description | Mannose-binding protein A precursor (MBP-A) (Mannan-binding protein) (Ra-reactive factor polysaccharide-binding component p28B polypeptide) (RaRF p28B). | |||||
|
MBL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.051838 (rank : 144) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11226, Q86SI4, Q96KE4, Q96TF7, Q96TF8, Q96TF9 | Gene names | MBL2, MBL | |||
|
Domain Architecture |
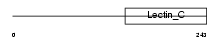
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (MBP1) (Mannan-binding protein) (Mannose-binding lectin). | |||||
|
MBL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.058515 (rank : 125) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |
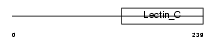
|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
MMGL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053223 (rank : 139) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |
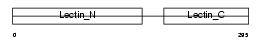
|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
MMP10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.075216 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.075984 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |
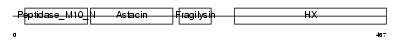
|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.076750 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.077156 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
MMP12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.071498 (rank : 69) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
MMP12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.073439 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.077210 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.076330 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.068081 (rank : 82) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
MMP14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.067794 (rank : 84) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
MMP15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.063185 (rank : 107) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
MMP15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.060941 (rank : 116) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
MMP16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.067490 (rank : 89) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
MMP16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.067357 (rank : 91) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
MMP17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.069913 (rank : 77) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.070194 (rank : 73) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
MMP19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.064434 (rank : 100) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |
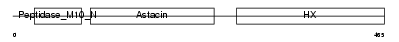
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
MMP19_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.070003 (rank : 75) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |
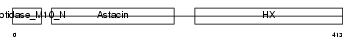
|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.074235 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
MMP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.072394 (rank : 66) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
MMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.075098 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
MMP20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.072747 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.072797 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.058426 (rank : 126) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.055889 (rank : 131) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
MMP24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.067529 (rank : 87) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
MMP24_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.068370 (rank : 81) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.069929 (rank : 76) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.079927 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |
|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
MMP28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.061877 (rank : 114) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
MMP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.074867 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.074729 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.086515 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.086536 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.071139 (rank : 71) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
MMP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.072653 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
NKG2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052224 (rank : 142) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26715 | Gene names | KLRC1, NKG2A | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-A/NKG2-B type II integral membrane protein (NKG2-A/B-activating NK receptor) (NK cell receptor A) (CD159a antigen). | |||||
|
NKG2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050303 (rank : 148) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26717, O43802, Q9NR42 | Gene names | KLRC2, NKG2C | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-C type II integral membrane protein (NKG2-C-activating NK receptor) (NK cell receptor C) (CD159c antigen). | |||||
|
NKG2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.060900 (rank : 119) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26718, Q9NR41 | Gene names | KLRK1, NKG2D | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
NKG2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.062578 (rank : 111) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54709 | Gene names | Klrk1, Nkg2d | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
NKG2E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051145 (rank : 146) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07444, Q96RL0, Q9UP04 | Gene names | KLRC3, NKG2E | |||
|
Domain Architecture |
|
|||||
| Description | NKG2-E type II integral membrane protein (NKG2-E-activating NK receptor) (NK cell receptor E). | |||||
|
OLR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052533 (rank : 141) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78380, Q2PP00, Q7Z484 | Gene names | OLR1, LOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) (hLOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
PGCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.064007 (rank : 104) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PGCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.064357 (rank : 101) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.072428 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
PGCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.071227 (rank : 70) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PRG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.091753 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
PRG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.087439 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
REG1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.079741 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |
|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
REG1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.077992 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |
|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
REG3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.073951 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
REG3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.083502 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09037 | Gene names | Reg3a, Pap2 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 2) (Lithostathine 3) (Islet of Langerhans regenerating protein 3). | |||||
|
REG3B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.079124 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
REG3G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.075248 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UW15, Q6FH18 | Gene names | REG3G, PAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB). | |||||
|
REG3G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.081967 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |
|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
REG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.076108 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
REG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.084065 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
SFTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.056091 (rank : 130) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWL2, P07714, Q5RIR5, Q5RIR7, Q6PIT0, Q8TC19 | Gene names | SFTPA1, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A1 precursor (SP-A1) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
SFTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.055449 (rank : 132) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWL1, P07714, Q5RIR8, Q5RIR9 | Gene names | SFTPA2, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A2 precursor (SP-A2) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
TETN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.077029 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
TETN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.077787 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43025 | Gene names | Clec3b, Tna | |||
|
Domain Architecture |
|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
VTNC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.051355 (rank : 145) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.994744 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MPRI_MOUSE
|
||||||
| NC score | 0.442330 (rank : 3) | θ value | 9.59137e-10 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q07113, Q61822, Q6LED1 | Gene names | Igf2r | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300). | |||||
|
MPRI_HUMAN
|
||||||
| NC score | 0.433332 (rank : 4) | θ value | 3.64472e-09 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P11717, Q7Z7G9, Q96PT5 | Gene names | IGF2R, MPRI | |||
|
Domain Architecture |

|
|||||
| Description | Cation-independent mannose-6-phosphate receptor precursor (CI Man-6-P receptor) (CI-MPR) (M6PR) (Insulin-like growth factor 2 receptor) (Insulin-like growth factor II receptor) (IGF-II receptor) (M6P/IGF2 receptor) (M6P/IGF2R) (300 kDa mannose 6-phosphate receptor) (MPR 300) (MPR300) (CD222 antigen). | |||||
|
MRC2_MOUSE
|
||||||
| NC score | 0.403567 (rank : 5) | θ value | 4.91457e-14 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64449, Q6ZQ64, Q8C6P0 | Gene names | Mrc2, Kiaa0709 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Lectin lambda) (CD280 antigen). | |||||
|
MRC1_MOUSE
|
||||||
| NC score | 0.402620 (rank : 6) | θ value | 6.64225e-11 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61830, Q8C502 | Gene names | Mrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MRC2_HUMAN
|
||||||
| NC score | 0.401992 (rank : 7) | θ value | 4.91457e-14 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBG0, Q7LGE7, Q9Y5P9 | Gene names | MRC2, ENDO180, KIAA0709, UPARAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macrophage mannose receptor 2 precursor (Urokinase receptor-associated protein) (Endocytic receptor 180) (CD280 antigen). | |||||
|
MMP9_MOUSE
|
||||||
| NC score | 0.394510 (rank : 8) | θ value | 2.35696e-16 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P41245, Q06788, Q9DC02 | Gene names | Mmp9, Clg4b | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.393023 (rank : 9) | θ value | 6.64225e-11 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
MMP9_HUMAN
|
||||||
| NC score | 0.390595 (rank : 10) | θ value | 5.8054e-15 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P14780, Q3LR70, Q8N725, Q9H4Z1 | Gene names | MMP9, CLG4B | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-9 precursor (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB) [Contains: 67 kDa matrix metalloproteinase-9; 82 kDa matrix metalloproteinase-9]. | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.384626 (rank : 11) | θ value | 3.08544e-08 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
LY75_HUMAN
|
||||||
| NC score | 0.378274 (rank : 12) | θ value | 3.41135e-07 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60449, O75913, Q7Z575, Q7Z577 | Gene names | LY75, CD205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (gp200-MR6) (CD205 antigen). | |||||
|
K0141_MOUSE
|
||||||
| NC score | 0.366345 (rank : 13) | θ value | 3.29651e-10 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCV6, Q6A0B8 | Gene names | Kiaa0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
K0141_HUMAN
|
||||||
| NC score | 0.357557 (rank : 14) | θ value | 5.62301e-10 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14154, Q969R4, Q96EU9 | Gene names | KIAA0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
MMP2_HUMAN
|
||||||
| NC score | 0.354355 (rank : 15) | θ value | 2.88119e-14 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08253 | Gene names | MMP2, CLG4A | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A) (TBE- 1). | |||||
|
MMP2_MOUSE
|
||||||
| NC score | 0.353886 (rank : 16) | θ value | 3.76295e-14 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33434 | Gene names | Mmp2 | |||
|
Domain Architecture |

|
|||||
| Description | 72 kDa type IV collagenase precursor (EC 3.4.24.24) (72 kDa gelatinase) (Matrix metalloproteinase-2) (MMP-2) (Gelatinase A). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.296858 (rank : 17) | θ value | 1.74796e-11 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.296624 (rank : 18) | θ value | 3.89403e-11 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
EF2K_MOUSE
|
||||||
| NC score | 0.186391 (rank : 19) | θ value | 0.000270298 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| NC score | 0.176522 (rank : 20) | θ value | 0.000786445 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.148875 (rank : 21) | θ value | 2.79066e-09 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.140975 (rank : 22) | θ value | 1.17247e-07 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.137589 (rank : 23) | θ value | 5.81887e-07 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
CD302_HUMAN
|
||||||
| NC score | 0.125616 (rank : 24) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX05, Q15009 | Gene names | CD302, CLEC13A, DCL1, KIAA0022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (C- type lectin BIMLEC) (Type I transmembrane C-type lectin receptor DCL- 1). | |||||
|
CD302_MOUSE
|
||||||
| NC score | 0.125058 (rank : 25) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DCG2, Q78KD8, Q9D0X7 | Gene names | Cd302, Clec13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD302 antigen precursor (C-type lectin domain family 13 member A) (Type I transmembrane C-type lectin receptor DCL-1). | |||||
|
CHODL_HUMAN
|
||||||
| NC score | 0.101530 (rank : 26) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H9P2, Q7Z798, Q7Z799, Q7Z7A0, Q9HCY3 | Gene names | CHODL, C21orf68 | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
CHODL_MOUSE
|
||||||
| NC score | 0.100867 (rank : 27) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXM0, Q8VI31 | Gene names | Chodl | |||
|
Domain Architecture |

|
|||||
| Description | Chondrolectin precursor (Transmembrane protein MT75). | |||||
|
PRG3_HUMAN
|
||||||
| NC score | 0.091753 (rank : 28) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2Y8 | Gene names | PRG3, MBPH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein homolog) (Prepro-major basic protein homolog) (Prepro-MBPH). | |||||
|
EMBP_MOUSE
|
||||||
| NC score | 0.087991 (rank : 29) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
PRG3_MOUSE
|
||||||
| NC score | 0.087439 (rank : 30) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
MMP7_MOUSE
|
||||||
| NC score | 0.086536 (rank : 31) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q10738 | Gene names | Mmp7 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
MMP7_HUMAN
|
||||||
| NC score | 0.086515 (rank : 32) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09237, Q9BTK9 | Gene names | MMP7, MPSL1, PUMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilysin precursor (EC 3.4.24.23) (Pump-1 protease) (Uterine metalloproteinase) (Matrix metalloproteinase-7) (MMP-7) (Matrin). | |||||
|
REG4_MOUSE
|
||||||
| NC score | 0.084065 (rank : 33) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
REG3A_MOUSE
|
||||||
| NC score | 0.083502 (rank : 34) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09037 | Gene names | Reg3a, Pap2 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 2) (Lithostathine 3) (Islet of Langerhans regenerating protein 3). | |||||
|
REG3G_MOUSE
|
||||||
| NC score | 0.081967 (rank : 35) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09049 | Gene names | Reg3g, Pap3 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 3). | |||||
|
EMBP_HUMAN
|
||||||
| NC score | 0.081212 (rank : 36) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P13727, Q14227 | Gene names | PRG2, MBP | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (EMBP) (Pregnancy-associated major basic protein) (Proteoglycan 2, bone marrow) (BMPG). | |||||
|
MMP26_HUMAN
|
||||||
| NC score | 0.079927 (rank : 37) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRE1, Q9GZS2, Q9NR87 | Gene names | MMP26 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-26 precursor (EC 3.4.24.-) (MMP-26) (Matrilysin-2) (Endometase). | |||||
|
REG1A_HUMAN
|
||||||
| NC score | 0.079741 (rank : 38) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05451, P11379 | Gene names | REG1A, PSPS, REG | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 alpha precursor (Pancreatic stone protein) (PSP) (Pancreatic thread protein) (PTP) (Islet of Langerhans regenerating protein) (REG) (Regenerating protein I alpha) (Islet cells regeneration factor) (ICRF). | |||||
|
FCER2_HUMAN
|
||||||
| NC score | 0.079238 (rank : 39) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06734 | Gene names | FCER2, IGEBF | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (BLAST-2) (Immunoglobulin E-binding factor) (CD23 antigen) [Contains: Low affinity immunoglobulin epsilon Fc receptor membrane-bound form; Low affinity immunoglobulin epsilon Fc receptor soluble form]. | |||||
|
REG3B_MOUSE
|
||||||
| NC score | 0.079124 (rank : 40) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35230 | Gene names | Reg3b, Pap, Pap1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 beta precursor (Reg III-beta) (Pancreatitis-associated protein 1). | |||||
|
REG1B_HUMAN
|
||||||
| NC score | 0.077992 (rank : 41) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P48304 | Gene names | REG1B, REGL | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 beta precursor (Regenerating protein I beta). | |||||
|
TETN_MOUSE
|
||||||
| NC score | 0.077787 (rank : 42) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43025 | Gene names | Clec3b, Tna | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
MMP13_HUMAN
|
||||||
| NC score | 0.077210 (rank : 43) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P45452 | Gene names | MMP13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.077156 (rank : 44) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
TETN_HUMAN
|
||||||
| NC score | 0.077029 (rank : 45) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P05452 | Gene names | CLEC3B, TNA | |||
|
Domain Architecture |

|
|||||
| Description | Tetranectin precursor (TN) (C-type lectin domain family 3 member B) (Plasminogen kringle 4-binding protein). | |||||
|
MMP11_HUMAN
|
||||||
| NC score | 0.076750 (rank : 46) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24347, Q5FX24, Q6PEZ6, Q9UC26 | Gene names | MMP11, STMY3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
LIT2_MOUSE
|
||||||
| NC score | 0.076651 (rank : 47) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08731 | Gene names | Reg2 | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 2 precursor (Pancreatic stone protein 2) (PSP) (Pancreatic thread protein 2) (PTP) (Islet of Langerhans regenerating protein 2) (REG 2). | |||||
|
MMP13_MOUSE
|
||||||
| NC score | 0.076330 (rank : 48) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33435 | Gene names | Mmp13 | |||
|
Domain Architecture |

|
|||||
| Description | Collagenase 3 precursor (EC 3.4.24.-) (Matrix metalloproteinase-13) (MMP-13). | |||||
|
CLC4G_MOUSE
|
||||||
| NC score | 0.076279 (rank : 49) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BNX1, Q5I0T7, Q8R188 | Gene names | Clec4g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
FCER2_MOUSE
|
||||||
| NC score | 0.076108 (rank : 50) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20693, Q61556, Q61557 | Gene names | Fcer2, Fcer2a | |||
|
Domain Architecture |

|
|||||
| Description | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IGE receptor) (Fc-epsilon-RII) (CD23 antigen). | |||||
|
REG4_HUMAN
|
||||||
| NC score | 0.076108 (rank : 51) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
MMP10_MOUSE
|
||||||
| NC score | 0.075984 (rank : 52) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55123 | Gene names | Mmp10 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
REG3G_HUMAN
|
||||||
| NC score | 0.075248 (rank : 53) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UW15, Q6FH18 | Gene names | REG3G, PAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 3 gamma precursor (Reg III-gamma) (Pancreatitis-associated protein 1B) (PAP IB). | |||||
|
MMP10_HUMAN
|
||||||
| NC score | 0.075216 (rank : 54) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09238 | Gene names | MMP10, STMY2 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-2 precursor (EC 3.4.24.22) (Matrix metalloproteinase-10) (MMP-10) (Transin-2) (SL-2). | |||||
|
MMP1_HUMAN
|
||||||
| NC score | 0.075098 (rank : 55) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P03956, P08156 | Gene names | MMP1, CLG | |||
|
Domain Architecture |

|
|||||
| Description | Interstitial collagenase precursor (EC 3.4.24.7) (Matrix metalloproteinase-1) (MMP-1) (Fibroblast collagenase) [Contains: 22 kDa interstitial collagenase; 27 kDa interstitial collagenase]. | |||||
|
KLRF1_HUMAN
|
||||||
| NC score | 0.075070 (rank : 56) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZS2, Q96PR2, Q96PR3, Q9NZS1 | Gene names | KLRF1, ML | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor subfamily F member 1 (Lectin-like receptor F1) (Activating coreceptor NKp80). | |||||
|
MMP3_HUMAN
|
||||||
| NC score | 0.074867 (rank : 57) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08254, Q3B7S0 | Gene names | MMP3, STMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1). | |||||
|
MMP3_MOUSE
|
||||||
| NC score | 0.074729 (rank : 58) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28862 | Gene names | Mmp3 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-1 precursor (EC 3.4.24.17) (Matrix metalloproteinase-3) (MMP-3) (Transin-1) (SL-1) (EMS-2). | |||||
|
MMP1A_MOUSE
|
||||||
| NC score | 0.074235 (rank : 59) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPL5 | Gene names | Mmp1a, McolA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase A precursor (EC 3.4.24.7) (Matrix metalloproteinase-1a) (MMP-1a) (Mcol-A). | |||||
|
REG3A_HUMAN
|
||||||
| NC score | 0.073951 (rank : 60) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06141 | Gene names | REG3A, HIP, PAP, PAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regenerating islet-derived protein 3 alpha precursor (Reg III-alpha) (Pancreatitis-associated protein 1). | |||||
|
MMP12_MOUSE
|
||||||
| NC score | 0.073439 (rank : 61) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P34960, Q8BJ92, Q8BJB3, Q8BJB6, Q8BJB8, Q8BJB9, Q8VED6 | Gene names | Mmp12, Mme, Mmel | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (MME) (Matrix metalloproteinase-12) (MMP-12). | |||||
|
MMP20_MOUSE
|
||||||
| NC score | 0.072797 (rank : 62) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57748 | Gene names | Mmp20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP20_HUMAN
|
||||||
| NC score | 0.072747 (rank : 63) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60882, Q6DKT9 | Gene names | MMP20 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-20 precursor (EC 3.4.24.-) (MMP-20) (Enamel metalloproteinase) (Enamelysin). | |||||
|
MMP8_MOUSE
|
||||||
| NC score | 0.072653 (rank : 64) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70138, O88733 | Gene names | Mmp8 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (Collagenase 2). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.072428 (rank : 65) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
MMP1B_MOUSE
|
||||||
| NC score | 0.072394 (rank : 66) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EPL6 | Gene names | Mmp1b, McolB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interstitial collagenase B precursor (EC 3.4.24.7) (Matrix metalloproteinase-1b) (MMP-1b) (Mcol-B). | |||||
|
CLC4G_HUMAN
|
||||||
| NC score | 0.072296 (rank : 67) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UXB4 | Gene names | CLEC4G | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member G. | |||||
|
LIT1_MOUSE
|
||||||
| NC score | 0.071674 (rank : 68) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43137 | Gene names | Reg1 | |||
|
Domain Architecture |

|
|||||
| Description | Lithostathine 1 precursor (Pancreatic stone protein 1) (PSP) (Pancreatic thread protein 1) (PTP) (Islet of Langerhans regenerating protein 1) (REG 1). | |||||
|
MMP12_HUMAN
|
||||||
| NC score | 0.071498 (rank : 69) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P39900 | Gene names | MMP12, HME | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage metalloelastase precursor (EC 3.4.24.65) (HME) (Matrix metalloproteinase-12) (MMP-12) (Macrophage elastase) (ME). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.071227 (rank : 70) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
MMP8_HUMAN
|
||||||
| NC score | 0.071139 (rank : 71) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22894, Q45F99 | Gene names | MMP8, CLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil collagenase precursor (EC 3.4.24.34) (Matrix metalloproteinase-8) (MMP-8) (PMNL collagenase) (PMNL-CL). | |||||
|
CLC11_HUMAN
|
||||||
| NC score | 0.070694 (rank : 72) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y240 | Gene names | CLEC11A, CLECSF3, LSLCL, SCGF | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin) (p47) (C-type lectin superfamily member 3). | |||||
|
MMP17_MOUSE
|
||||||
| NC score | 0.070194 (rank : 73) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
KLRD1_HUMAN
|
||||||
| NC score | 0.070099 (rank : 74) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13241, O43321, O43773, Q9UBE3, Q9UEQ0 | Gene names | KLRD1, CD94 | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cells antigen CD94 (NK cell receptor) (Killer cell lectin-like receptor subfamily D member 1) (KP43). | |||||
|
MMP19_MOUSE
|
||||||
| NC score | 0.070003 (rank : 75) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHI0, Q8C360 | Gene names | Mmp19, Rasi | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI). | |||||
|
MMP25_HUMAN
|
||||||
| NC score | 0.069929 (rank : 76) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NPA2, Q9H3Q0 | Gene names | MMP25, MT6MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-25 precursor (EC 3.4.24.-) (MMP-25) (Membrane-type matrix metalloproteinase 6) (MT-MMP 6) (Membrane-type-6 matrix metalloproteinase) (MT6-MMP) (Leukolysin). | |||||
|
MMP17_HUMAN
|
||||||
| NC score | 0.069913 (rank : 77) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULZ9, Q14850 | Gene names | MMP17, MT4MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.069138 (rank : 78) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CLC4E_HUMAN
|
||||||
| NC score | 0.068592 (rank : 79) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULY5 | Gene names | CLEC4E, CLECSF9, MINCLE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
CLC4D_MOUSE
|
||||||
| NC score | 0.068523 (rank : 80) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2H6, Q8C212 | Gene names | Clec4d, Clecsf8, Mcl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (Macrophage-restricted C-type lectin). | |||||
|
MMP24_MOUSE
|
||||||
| NC score | 0.068370 (rank : 81) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R0S2, Q9Z0J9 | Gene names | Mmp24, Mmp21, Mt5mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21). | |||||
|
MMP14_HUMAN
|
||||||
| NC score | 0.068081 (rank : 82) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50281, Q92678 | Gene names | MMP14 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP- X1). | |||||
|
CLC4A_MOUSE
|
||||||
| NC score | 0.067926 (rank : 83) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZ15, Q4VA33 | Gene names | Clec4a, Clec4a2, Clecsf6, Dcir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor). | |||||
|
MMP14_MOUSE
|
||||||
| NC score | 0.067794 (rank : 84) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53690, O08645, O35369 | Gene names | Mmp14, Mtmmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-14 precursor (EC 3.4.24.80) (MMP-14) (Membrane-type matrix metalloproteinase 1) (MT-MMP 1) (MTMMP1) (Membrane-type-1 matrix metalloproteinase) (MT1-MMP) (MT1MMP) (MMP-X1) (MT-MMP). | |||||
|
CLC6A_HUMAN
|
||||||
| NC score | 0.067718 (rank : 85) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.067538 (rank : 86) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
MMP24_HUMAN
|
||||||
| NC score | 0.067529 (rank : 87) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5R2, Q9H440 | Gene names | MMP24, MT5MMP | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-24 precursor (EC 3.4.24.-) (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP). | |||||
|
C209D_MOUSE
|
||||||
| NC score | 0.067519 (rank : 88) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZW8, Q8VIK4 | Gene names | Cd209d | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein D (DC-SIGN-related protein 3) (DC-SIGNR3). | |||||
|
MMP16_HUMAN
|
||||||
| NC score | 0.067490 (rank : 89) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51512, Q14824, Q52H48 | Gene names | MMP16, MMPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP) (MMP- X2). | |||||
|
CLC4E_MOUSE
|
||||||
| NC score | 0.067373 (rank : 90) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0Q8 | Gene names | Clec4e, Clecsf9, Mincle | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member E (C-type lectin superfamily member 9) (Macrophage-inducible C-type lectin). | |||||
|
MMP16_MOUSE
|
||||||
| NC score | 0.067357 (rank : 91) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTR0, Q9ERT6 | Gene names | Mmp16 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-16 precursor (EC 3.4.24.-) (MMP-16) (Membrane-type matrix metalloproteinase 3) (MT-MMP 3) (MTMMP3) (Membrane-type-3 matrix metalloproteinase) (MT3-MMP) (MT3MMP). | |||||
|
CLC3A_MOUSE
|
||||||
| NC score | 0.067204 (rank : 92) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPW4 | Gene names | Clec3a, Clecsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.067179 (rank : 93) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CLC4A_HUMAN
|
||||||
| NC score | 0.066683 (rank : 94) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UMR7, Q8WXW9, Q9H2Z9, Q9NS33, Q9UI34 | Gene names | CLEC4A, CLECSF6, DCIR, LLIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member A (C-type lectin superfamily member 6) (Dendritic cell immunoreceptor) (Lectin-like immunoreceptor) (C-type lectin DDB27). | |||||
|
CLC6A_MOUSE
|
||||||
| NC score | 0.066535 (rank : 95) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKF4, Q9JKF2, Q9JKF3 | Gene names | Clec6a, Clecsf10, Dectin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
CLC3A_HUMAN
|
||||||
| NC score | 0.065046 (rank : 96) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75596, Q6UXF5 | Gene names | CLEC3A, CLECSF1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 3 member A precursor (C-type lectin superfamily member 1) (Cartilage-derived C-type lectin). | |||||
|
CLC4K_HUMAN
|
||||||
| NC score | 0.064966 (rank : 97) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJ71 | Gene names | CD207, CLEC4K | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC4C_HUMAN
|
||||||
| NC score | 0.064884 (rank : 98) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WTT0, Q3T1C3, Q6UXS8, Q8WXX8 | Gene names | CLEC4C, BDCA2, CLECSF11, CLECSF7, DLEC, HECL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member C (C-type lectin superfamily member 7) (Blood dendritic cell antigen 2 protein) (BDCA-2) (Dendritic lectin) (CD303 antigen). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.064640 (rank : 99) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MMP19_HUMAN
|
||||||
| NC score | 0.064434 (rank : 100) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99542, O15278, O95606, Q99580 | Gene names | MMP19, MMP18, RASI | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-19 precursor (EC 3.4.24.-) (MMP-19) (Matrix metalloproteinase RASI) (MMP-18). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.064357 (rank : 101) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
C209C_MOUSE
|
||||||
| NC score | 0.064225 (rank : 102) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZW9 | Gene names | Cd209c | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein C (DC-SIGN-related protein 2) (DC-SIGNR2). | |||||
|
WFS1_HUMAN
|
||||||
| NC score | 0.064115 (rank : 103) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76024, Q8N6I3, Q9UNW6 | Gene names | WFS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wolframin. | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.064007 (rank : 104) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CLC11_MOUSE
|
||||||
| NC score | 0.063789 (rank : 105) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88200, Q8C946, Q9CTF0 | Gene names | Clec11a, Scgf | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 11 member A precursor (Stem cell growth factor) (Lymphocyte secreted C-type lectin). | |||||
|
C209E_MOUSE
|
||||||
| NC score | 0.063484 (rank : 106) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZW7 | Gene names | Cd209e | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein E (DC-SIGN-related protein 4) (DC-SIGNR4). | |||||
|
MMP15_HUMAN
|
||||||
| NC score | 0.063185 (rank : 107) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51511, Q14111 | Gene names | MMP15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP) (SMCP- 2). | |||||
|
CLC4D_HUMAN
|
||||||
| NC score | 0.063046 (rank : 108) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
CD209_HUMAN
|
||||||
| NC score | 0.062862 (rank : 109) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
C209B_MOUSE
|
||||||
| NC score | 0.062623 (rank : 110) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CJ91, Q8BGZ0, Q8BHK7, Q8CJ86, Q8CJ87, Q8CJ88, Q8CJ89, Q8CJ90, Q8CJ92, Q8CJ93, Q8CJ94, Q91ZW4, Q91ZX0, Q9D8V4 | Gene names | Cd209b | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein B (DC-SIGN-related protein 1) (DC-SIGNR1) (OtB7). | |||||
|
NKG2D_MOUSE
|
||||||
| NC score | 0.062578 (rank : 111) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54709 | Gene names | Klrk1, Nkg2d | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
C209A_MOUSE
|
||||||
| NC score | 0.062179 (rank : 112) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZX1, Q3TAT9, Q91ZW5 | Gene names | Cd209a, Cire | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen-like protein A (Dendritic cell-specific ICAM-3-grabbing nonintegrin) (DC-SIGN). | |||||
|
KLRBB_MOUSE
|
||||||
| NC score | 0.061963 (rank : 113) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27812 | Gene names | Klrb1b, Ly55-b, Ly55b | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell surface protein P1-34 (NKR-P1 34). | |||||
|
MMP28_HUMAN
|
||||||
| NC score | 0.061877 (rank : 114) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H239, Q96TE2 | Gene names | MMP28, MMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-28 precursor (EC 3.4.24.-) (MMP-28) (Epilysin). | |||||
|
CLC4M_HUMAN
|
||||||
| NC score | 0.060986 (rank : 115) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
MMP15_MOUSE
|
||||||
| NC score | 0.060941 (rank : 116) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54732 | Gene names | Mmp15 | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-15 precursor (EC 3.4.24.-) (MMP-15) (Membrane-type matrix metalloproteinase 2) (MT-MMP 2) (MTMMP2) (Membrane-type-2 matrix metalloproteinase) (MT2-MMP) (MT2MMP). | |||||
|
CD69_HUMAN
|
||||||
| NC score | 0.060933 (rank : 117) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |

|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
CLC9A_HUMAN
|
||||||
| NC score | 0.060926 (rank : 118) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UXN8 | Gene names | CLEC9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
NKG2D_HUMAN
|
||||||
| NC score | 0.060900 (rank : 119) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26718, Q9NR41 | Gene names | KLRK1, NKG2D | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-D type II integral membrane protein (NKG2-D-activating NK receptor) (NK cell receptor D) (Killer cell lectin-like receptor subfamily K member 1) (CD314 antigen). | |||||
|
CLC4K_MOUSE
|
||||||
| NC score | 0.060898 (rank : 120) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VBX4, Q8R442 | Gene names | Cd207, Clec4k | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member K (Langerin) (CD207 antigen). | |||||
|
CLC1A_MOUSE
|
||||||
| NC score | 0.060559 (rank : 121) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BWY2, Q7TPX7, Q8BMH5 | Gene names | Clec1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A. | |||||
|
ASGR2_MOUSE
|
||||||
| NC score | 0.060360 (rank : 122) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24721 | Gene names | Asgr2, Asgr-2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin 2) (MHL-2). | |||||
|
KLRBA_MOUSE
|
||||||
| NC score | 0.060353 (rank : 123) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27811 | Gene names | Klrb1a, Ly55, Ly55a | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell surface protein P1-2 (NKR-P1 2) (NKR-P1.7). | |||||
|
ASGR2_HUMAN
|
||||||
| NC score | 0.058655 (rank : 124) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P07307, O00448, Q03969 | Gene names | ASGR2 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 2 (ASGP-R 2) (ASGPR 2) (Hepatic lectin H2). | |||||
|
MBL2_MOUSE
|
||||||
| NC score | 0.058515 (rank : 125) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41317 | Gene names | Mbl2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (Mannan-binding protein) (RA-reactive factor P28A subunit) (RARF/P28A). | |||||
|
MMP21_HUMAN
|
||||||
| NC score | 0.058426 (rank : 126) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N119, Q8NG02 | Gene names | MMP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
KLRBC_MOUSE
|
||||||
| NC score | 0.058177 (rank : 127) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27814 | Gene names | Klrb1c, Ly55-c, Ly55c | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell surface protein P1-40 (NKR-P1 40) (NKR-P1.9). | |||||
|
CLC10_HUMAN
|
||||||
| NC score | 0.057355 (rank : 128) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
CD69_MOUSE
|
||||||
| NC score | 0.056115 (rank : 129) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P37217 | Gene names | Cd69 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early activation antigen CD69. | |||||
|
SFTA1_HUMAN
|
||||||
| NC score | 0.056091 (rank : 130) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWL2, P07714, Q5RIR5, Q5RIR7, Q6PIT0, Q8TC19 | Gene names | SFTPA1, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A1 precursor (SP-A1) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
MMP21_MOUSE
|
||||||
| NC score | 0.055889 (rank : 131) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3F2 | Gene names | Mmp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrix metalloproteinase-21 precursor (EC 3.4.24.-) (MMP-21). | |||||
|
SFTA2_HUMAN
|
||||||
| NC score | 0.055449 (rank : 132) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWL1, P07714, Q5RIR8, Q5RIR9 | Gene names | SFTPA2, PSAP, SFTP1, SFTPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pulmonary surfactant-associated protein A2 precursor (SP-A2) (SP-A) (PSP-A) (PSPA) (Alveolar proteinosis protein) (35 kDa pulmonary surfactant-associated protein). | |||||
|
CLC1A_HUMAN
|
||||||
| NC score | 0.054558 (rank : 133) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NC01, Q8IUW7, Q9NZH3 | Gene names | CLEC1A, CLEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 1 member A (C-type lectin-like receptor 1) (CLEC-1). | |||||
|
LYAM1_HUMAN
|
||||||
| NC score | 0.054406 (rank : 134) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14151, P15023 | Gene names | SELL, LNHR, LYAM1 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Leukocyte surface antigen Leu-8) (TQ1) (gp90-MEL) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
HEMO_MOUSE
|
||||||
| NC score | 0.054391 (rank : 135) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91X72, P97824, Q8WUP0 | Gene names | Hpx, Hpxn | |||
|
Domain Architecture |

|
|||||
| Description | Hemopexin precursor. | |||||
|
LYAM1_MOUSE
|
||||||
| NC score | 0.054256 (rank : 136) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18337 | Gene names | Sell, Lnhr, Ly-22 | |||
|
Domain Architecture |

|
|||||
| Description | L-selectin precursor (Lymph node homing receptor) (Leukocyte adhesion molecule 1) (LAM-1) (Lymphocyte antigen 22) (Ly-22) (Lymphocyte surface MEL-14 antigen) (Leukocyte-endothelial cell adhesion molecule 1) (LECAM1) (CD62L antigen). | |||||
|
CLC9A_MOUSE
|
||||||
| NC score | 0.053965 (rank : 137) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRU4 | Gene names | Clec9a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 9 member A. | |||||
|
ASGR1_MOUSE
|
||||||
| NC score | 0.053695 (rank : 138) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
MMGL_MOUSE
|
||||||
| NC score | 0.053223 (rank : 139) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49300 | Gene names | Mgl1, Mgl | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage asialoglycoprotein-binding protein 1 (M-ASGP-BP) (Macrophage galactose/N-acetylgalactosamine-specific lectin) (MMGL). | |||||
|
MBL1_MOUSE
|
||||||
| NC score | 0.052548 (rank : 140) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39039 | Gene names | Mbl1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein A precursor (MBP-A) (Mannan-binding protein) (Ra-reactive factor polysaccharide-binding component p28B polypeptide) (RaRF p28B). | |||||
|
OLR1_HUMAN
|
||||||
| NC score | 0.052533 (rank : 141) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78380, Q2PP00, Q7Z484 | Gene names | OLR1, LOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) (hLOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
NKG2A_HUMAN
|
||||||
| NC score | 0.052224 (rank : 142) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26715 | Gene names | KLRC1, NKG2A | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-A/NKG2-B type II integral membrane protein (NKG2-A/B-activating NK receptor) (NK cell receptor A) (CD159a antigen). | |||||
|
IDD_MOUSE
|
||||||
| NC score | 0.051906 (rank : 143) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98154, Q61844 | Gene names | Dgcr2, Dgsc, Idd, Sez-12, Sez12 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor (Seizure-related membrane-bound adhesion protein). | |||||
|
MBL2_HUMAN
|
||||||
| NC score | 0.051838 (rank : 144) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11226, Q86SI4, Q96KE4, Q96TF7, Q96TF8, Q96TF9 | Gene names | MBL2, MBL | |||
|
Domain Architecture |

|
|||||
| Description | Mannose-binding protein C precursor (MBP-C) (MBP1) (Mannan-binding protein) (Mannose-binding lectin). | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.051355 (rank : 145) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
NKG2E_HUMAN
|
||||||
| NC score | 0.051145 (rank : 146) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07444, Q96RL0, Q9UP04 | Gene names | KLRC3, NKG2E | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-E type II integral membrane protein (NKG2-E-activating NK receptor) (NK cell receptor E). | |||||
|
FREM1_MOUSE
|
||||||
| NC score | 0.050559 (rank : 147) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q684R7, Q5H8C2, Q5M7B3, Q8C732 | Gene names | Frem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 1 precursor (Protein QBRICK). | |||||
|
NKG2C_HUMAN
|
||||||
| NC score | 0.050303 (rank : 148) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P26717, O43802, Q9NR42 | Gene names | KLRC2, NKG2C | |||
|
Domain Architecture |

|
|||||
| Description | NKG2-C type II integral membrane protein (NKG2-C-activating NK receptor) (NK cell receptor C) (CD159c antigen). | |||||
|
BMS1_HUMAN
|
||||||
| NC score | 0.040882 (rank : 149) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.030331 (rank : 150) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.029759 (rank : 151) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.027054 (rank : 152) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.024054 (rank : 153) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.023760 (rank : 154) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.022767 (rank : 155) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
CCNF_MOUSE
|
||||||
| NC score | 0.022547 (rank : 156) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51944, Q60797, Q60799 | Gene names | Ccnf | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
CCNF_HUMAN
|
||||||
| NC score | 0.022367 (rank : 157) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41002, Q96EG9 | Gene names | CCNF | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-F. | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.021254 (rank : 158) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
TRUB2_HUMAN
|
||||||
| NC score | 0.020508 (rank : 159) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95900 | Gene names | TRUB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tRNA pseudouridine synthase 2 (EC 5.4.99.-). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.020076 (rank : 160) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.018218 (rank : 161) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.017985 (rank : 162) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.017900 (rank : 163) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.016800 (rank : 164) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CHIC1_HUMAN
|
||||||
| NC score | 0.016363 (rank : 165) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VXU3 | Gene names | CHIC1, BRX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich hydrophobic domain 1 protein (Brain X-linked protein). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.016139 (rank : 166) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.014876 (rank : 167) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
DLG2_MOUSE
|
||||||
| NC score | 0.013992 (rank : 168) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
DLG2_HUMAN
|
||||||
| NC score | 0.013874 (rank : 169) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.013683 (rank : 170) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.013584 (rank : 171) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.012271 (rank : 172) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
AN32A_MOUSE
|
||||||
| NC score | 0.010999 (rank : 173) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35381, P97437 | Gene names | Anp32a, Anp32, Lanp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein). | |||||
|
AN32C_MOUSE
|
||||||
| NC score | 0.010999 (rank : 174) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64G17 | Gene names | Anp32c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C. | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.010481 (rank : 175) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
HEPH_MOUSE
|
||||||
| NC score | 0.010459 (rank : 176) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0Z4, Q6ZQ65, Q80Y80, Q8C4S2 | Gene names | Heph, Kiaa0698 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hephaestin precursor (EC 1.-.-.-). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.008297 (rank : 177) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.006593 (rank : 178) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.005929 (rank : 179) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DDX24_HUMAN
|
||||||
| NC score | 0.004822 (rank : 180) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR7 | Gene names | DDX24 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX24 (EC 3.6.1.-) (DEAD box protein 24). | |||||
|
ABCB6_HUMAN
|
||||||
| NC score | 0.004733 (rank : 181) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP58, O75542 | Gene names | ABCB6, MTABC3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family B member 6, mitochondrial precursor (Mitochondrial ABC transporter 3) (Mt-ABC transporter 3) (ABC transporter umat). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.002209 (rank : 182) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.000543 (rank : 183) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MARK3_MOUSE
|
||||||
| NC score | -0.000606 (rank : 184) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03141, Q3TM40, Q3V1U3, Q8C6G9, Q8R375, Q9JKE4 | Gene names | Mark3, Emk2, Mpk10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP/microtubule affinity-regulating kinase 3 (EC 2.7.11.1) (MPK-10) (ELKL motif kinase 2). | |||||
|
KCC4_MOUSE
|
||||||
| NC score | -0.000653 (rank : 185) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08414, Q61381 | Gene names | Camk4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type IV (EC 2.7.11.17) (CAM kinase-GR) (CaMK IV). | |||||