Please be patient as the page loads
|
CAYP2_HUMAN
|
||||||
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAYP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.923933 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 3) | NC score | 0.570543 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
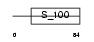
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 4) | NC score | 0.290728 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 5) | NC score | 0.301030 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
CETN2_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 6) | NC score | 0.251992 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
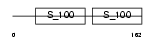
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 7) | NC score | 0.251512 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
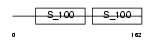
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 8) | NC score | 0.246474 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.233182 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.092823 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.237392 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
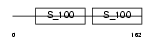
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.252926 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
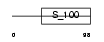
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.252984 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
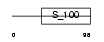
|
|||||
| Description | Calneuron-1. | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.190299 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
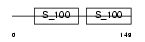
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.191007 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
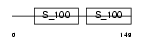
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.044881 (rank : 157) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.111031 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.110295 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.226894 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.167481 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.167472 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
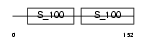
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
EFHB_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.136315 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.219526 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
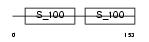
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CABP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.208195 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
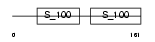
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.208247 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
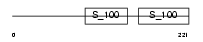
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.029612 (rank : 170) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
TNNT3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.050430 (rank : 155) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.090649 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.091453 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.093884 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.217143 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.091292 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MLRA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.163520 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |
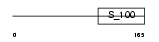
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.163137 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |
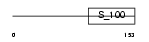
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.237943 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
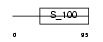
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.237943 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
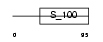
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.173073 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.038610 (rank : 159) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.219690 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.219690 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.199833 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
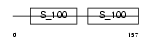
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
LZTS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.030216 (rank : 167) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.034513 (rank : 162) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
KI21B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.020274 (rank : 178) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
MLRS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.146701 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |
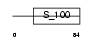
|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.020228 (rank : 179) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.200114 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
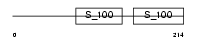
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.225638 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.037311 (rank : 160) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.020975 (rank : 176) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
ITIH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.022343 (rank : 174) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
PLCB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.032604 (rank : 163) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.030391 (rank : 166) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.030742 (rank : 165) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.026879 (rank : 172) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.225286 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.148179 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.022761 (rank : 173) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.017461 (rank : 180) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.028416 (rank : 171) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
ONCO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.147679 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |
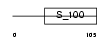
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
PPE2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.058201 (rank : 141) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.070380 (rank : 124) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.009864 (rank : 181) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
MCFD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.057786 (rank : 142) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NI22, Q8N3M5 | Gene names | MCFD2, SDNSF | |||
|
Domain Architecture |
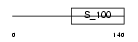
|
|||||
| Description | Multiple coagulation factor deficiency protein 2 precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
MYL6B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.160749 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
MYL6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.157141 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
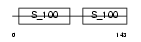
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.157141 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
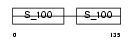
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
SEL1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.029895 (rank : 168) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.029759 (rank : 169) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
MLE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.143626 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.140665 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |
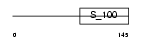
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
RYR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.035376 (rank : 161) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
ZBTB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.001528 (rank : 184) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2K1, Q86SW8 | Gene names | ZBTB1, KIAA0997 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||
|
CETN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.220244 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.220438 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
EFHB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.090508 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
GUC1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.084911 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
MCES_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.021690 (rank : 175) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
NCALD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.081656 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |
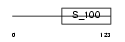
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.081656 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |
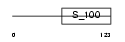
|
|||||
| Description | Neurocalcin delta. | |||||
|
NOC3L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.042524 (rank : 158) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
NOC3L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.031463 (rank : 164) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
ONCO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.143557 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |
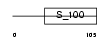
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.007709 (rank : 182) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.020401 (rank : 177) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
VISL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.075605 (rank : 113) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.075605 (rank : 114) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.094010 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.095899 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.192226 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.005672 (rank : 183) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
EFHC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.053701 (rank : 150) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D9T8 | Gene names | Efhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
GUC1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.077882 (rank : 108) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |
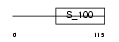
|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
MCFD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.049297 (rank : 156) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K5B2, Q3U9G5 | Gene names | Mcfd2, Sdnsf | |||
|
Domain Architecture |
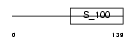
|
|||||
| Description | Multiple coagulation factor deficiency protein 2 homolog precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
MLE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.140190 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.136870 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |
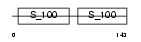
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
ZBTB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.001076 (rank : 185) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VL9, Q8CDP7, Q99LD2 | Gene names | Zbtb1 | |||
|
Domain Architecture |
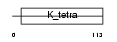
|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||
|
ACTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.087294 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.086324 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.085414 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
ACTN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.084379 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
AIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.067657 (rank : 128) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P55008, Q9UKS9 | Gene names | AIF1, G1, IBA1 | |||
|
Domain Architecture |
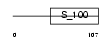
|
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1) (Protein G1). | |||||
|
AIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.067191 (rank : 129) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70200 | Gene names | Aif1, Iba1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1). | |||||
|
AYT1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.057397 (rank : 144) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.059861 (rank : 137) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.176273 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
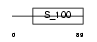
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.187264 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.194424 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.194296 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.078832 (rank : 105) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |
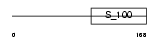
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CALB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.079536 (rank : 103) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |
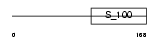
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.090756 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.093676 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.149904 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
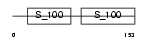
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CHP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.100817 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99653 | Gene names | CHP | |||
|
Domain Architecture |
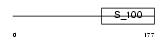
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog). | |||||
|
CHP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.102099 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |
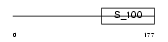
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
CHP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.102662 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
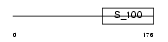
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
CHP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.104584 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |
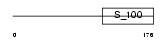
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
CPNS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051751 (rank : 153) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
CSEN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.055887 (rank : 145) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.051869 (rank : 152) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
DUOX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055269 (rank : 149) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
EFCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.069412 (rank : 126) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.075784 (rank : 112) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.143621 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFCB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.158829 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFCB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.073340 (rank : 120) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N7B9 | Gene names | EFCAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
EFCB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.076322 (rank : 111) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80X60 | Gene names | Efcab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
EFHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.099287 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |
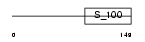
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
EFHD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.099435 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |
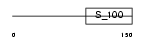
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
EFHD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.082533 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96C19 | Gene names | EFHD2, SWS1 | |||
|
Domain Architecture |
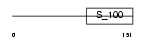
|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
EFHD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.083242 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D8Y0, Q9D0P4 | Gene names | Efhd2, Sws1 | |||
|
Domain Architecture |
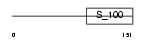
|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.080822 (rank : 99) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
GPDM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.078691 (rank : 106) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
GRAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.060948 (rank : 136) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |
|
|||||
| Description | Grancalcin. | |||||
|
GUC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.078888 (rank : 104) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.064950 (rank : 130) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.081953 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.073409 (rank : 118) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.073409 (rank : 119) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
HPCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.076545 (rank : 110) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.074045 (rank : 117) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.071451 (rank : 122) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.071449 (rank : 123) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
IBA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.062905 (rank : 133) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BQI0, Q6ZR40, Q8NAX7, Q8WU47, Q9H9G0 | Gene names | IBA2, C9orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
IBA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.063884 (rank : 131) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQX4, Q8C150 | Gene names | Iba2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
KCIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.055325 (rank : 147) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.055311 (rank : 148) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.074845 (rank : 115) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |
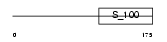
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
KIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.074379 (rank : 116) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |
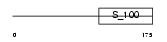
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
KIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.088650 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.087878 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.077101 (rank : 109) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96Q77 | Gene names | CIB3, KIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 3 (Kinase-interacting protein 3) (KIP 3). | |||||
|
MLR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.153223 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |
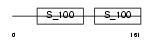
|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
MLRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.164472 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |
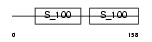
|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.163918 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.162763 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.144719 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.142724 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.129338 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
MYL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.132781 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MYL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.126045 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
MYL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.125854 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P09541 | Gene names | Myl4, Mlc1a, Myla | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, atrial/fetal isoform) (MLC1A) (MLC1EMB). | |||||
|
NCS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.058788 (rank : 139) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.058788 (rank : 140) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
PEF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.059109 (rank : 138) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.061272 (rank : 135) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.080608 (rank : 100) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
PLSL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.078421 (rank : 107) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.079646 (rank : 102) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.069491 (rank : 125) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PLST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.069012 (rank : 127) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
PRVA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.117191 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
PRVA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.131508 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |
|
|||||
| Description | Parvalbumin alpha. | |||||
|
RCN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.057520 (rank : 143) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |
|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
RCN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.062966 (rank : 132) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BP92, O70341 | Gene names | Rcn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-2 precursor (Taipoxin-associated calcium-binding protein 49) (TCBP-49). | |||||
|
RECO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.051464 (rank : 154) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
RECO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.055635 (rank : 146) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
SEGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.053495 (rank : 151) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
|
|||||
| Description | Secretagogin. | |||||
|
SORCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.071456 (rank : 121) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |
|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
SORCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.062539 (rank : 134) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.079956 (rank : 101) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.201281 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.199951 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP2_MOUSE
|
||||||
| NC score | 0.923933 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BUG5, Q8C1J6 | Gene names | Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.570543 (rank : 3) | θ value | 5.43371e-13 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.301030 (rank : 4) | θ value | 5.44631e-05 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.290728 (rank : 5) | θ value | 1.09739e-05 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.252984 (rank : 6) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.252926 (rank : 7) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.251992 (rank : 8) | θ value | 0.00035302 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.251512 (rank : 9) | θ value | 0.00228821 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.246474 (rank : 10) | θ value | 0.00228821 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.237943 (rank : 11) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.237943 (rank : 12) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.237392 (rank : 13) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.233182 (rank : 14) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.226894 (rank : 15) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.225638 (rank : 16) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.225286 (rank : 17) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.220438 (rank : 18) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.220244 (rank : 19) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.219690 (rank : 20) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.219690 (rank : 21) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.219526 (rank : 22) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.217143 (rank : 23) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.208247 (rank : 24) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.208195 (rank : 25) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.201281 (rank : 26) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.200114 (rank : 27) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.199951 (rank : 28) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.199833 (rank : 29) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.194424 (rank : 30) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.194296 (rank : 31) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.192226 (rank : 32) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.191007 (rank : 33) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.190299 (rank : 34) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.187264 (rank : 35) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.176273 (rank : 36) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.173073 (rank : 37) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.167481 (rank : 38) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.167472 (rank : 39) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
MLRM_HUMAN
|
||||||
| NC score | 0.164472 (rank : 40) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| NC score | 0.163918 (rank : 41) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRA_HUMAN
|
||||||
| NC score | 0.163520 (rank : 42) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_MOUSE
|
||||||
| NC score | 0.163137 (rank : 43) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRN_MOUSE
|
||||||
| NC score | 0.162763 (rank : 44) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MYL6B_MOUSE
|
||||||
| NC score | 0.160749 (rank : 45) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
EFCB2_MOUSE
|
||||||
| NC score | 0.158829 (rank : 46) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.157141 (rank : 47) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.157141 (rank : 48) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MLR5_HUMAN
|
||||||
| NC score | 0.153223 (rank : 49) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |

|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.149904 (rank : 50) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.148179 (rank : 51) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
ONCO_MOUSE
|
||||||
| NC score | 0.147679 (rank : 52) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
MLRS_MOUSE
|
||||||
| NC score | 0.146701 (rank : 53) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
MLRV_HUMAN
|
||||||
| NC score | 0.144719 (rank : 54) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLE1_MOUSE
|
||||||
| NC score | 0.143626 (rank : 55) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
EFCB2_HUMAN
|
||||||
| NC score | 0.143621 (rank : 56) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
ONCO_HUMAN
|
||||||
| NC score | 0.143557 (rank : 57) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
MLRV_MOUSE
|
||||||
| NC score | 0.142724 (rank : 58) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLE3_MOUSE
|
||||||
| NC score | 0.140665 (rank : 59) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE1_HUMAN
|
||||||
| NC score | 0.140190 (rank : 60) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_HUMAN
|
||||||
| NC score | 0.136870 (rank : 61) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
EFHB_HUMAN
|
||||||
| NC score | 0.136315 (rank : 62) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
MYL3_MOUSE
|
||||||
| NC score | 0.132781 (rank : 63) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
PRVA_MOUSE
|
||||||
| NC score | 0.131508 (rank : 64) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |

|
|||||
| Description | Parvalbumin alpha. | |||||
|
MYL3_HUMAN
|
||||||
| NC score | 0.129338 (rank : 65) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
MYL4_HUMAN
|
||||||
| NC score | 0.126045 (rank : 66) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
MYL4_MOUSE
|
||||||
| NC score | 0.125854 (rank : 67) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P09541 | Gene names | Myl4, Mlc1a, Myla | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, atrial/fetal isoform) (MLC1A) (MLC1EMB). | |||||
|
PRVA_HUMAN
|
||||||
| NC score | 0.117191 (rank : 68) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.111031 (rank : 69) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.110295 (rank : 70) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CHP2_MOUSE
|
||||||
| NC score | 0.104584 (rank : 71) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.102662 (rank : 72) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
CHP1_MOUSE
|
||||||
| NC score | 0.102099 (rank : 73) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
CHP1_HUMAN
|
||||||
| NC score | 0.100817 (rank : 74) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99653 | Gene names | CHP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog). | |||||
|
EFHD1_MOUSE
|
||||||
| NC score | 0.099435 (rank : 75) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
EFHD1_HUMAN
|
||||||
| NC score | 0.099287 (rank : 76) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.095899 (rank : 77) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.094010 (rank : 78) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.093884 (rank : 79) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.093676 (rank : 80) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.092823 (rank : 81) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.091453 (rank : 82) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.091292 (rank : 83) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.090756 (rank : 84) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.090649 (rank : 85) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.090508 (rank : 86) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
KIP2_HUMAN
|
||||||
| NC score | 0.088650 (rank : 87) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.087878 (rank : 88) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
ACTN1_HUMAN
|
||||||
| NC score | 0.087294 (rank : 89) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12814, Q9BTN1 | Gene names | ACTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN1_MOUSE
|
||||||
| NC score | 0.086324 (rank : 90) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TPR4 | Gene names | Actn1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-1 (Alpha-actinin cytoskeletal isoform) (Non-muscle alpha-actinin-1) (F-actin cross-linking protein). | |||||
|
ACTN4_HUMAN
|
||||||
| NC score | 0.085414 (rank : 91) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43707, O76048 | Gene names | ACTN4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
GUC1A_MOUSE
|
||||||
| NC score | 0.084911 (rank : 92) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
ACTN4_MOUSE
|
||||||
| NC score | 0.084379 (rank : 93) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57780 | Gene names | Actn4 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-4 (Non-muscle alpha-actinin 4) (F-actin cross-linking protein). | |||||
|
EFHD2_MOUSE
|
||||||
| NC score | 0.083242 (rank : 94) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D8Y0, Q9D0P4 | Gene names | Efhd2, Sws1 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
EFHD2_HUMAN
|
||||||
| NC score | 0.082533 (rank : 95) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96C19 | Gene names | EFHD2, SWS1 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.081953 (rank : 96) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
NCALD_HUMAN
|
||||||
| NC score | 0.081656 (rank : 97) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| NC score | 0.081656 (rank : 98) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.080822 (rank : 99) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.080608 (rank : 100) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.079956 (rank : 101) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.079646 (rank : 102) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
CALB1_MOUSE
|
||||||
| NC score | 0.079536 (rank : 103) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
GUC1A_HUMAN
|
||||||
| NC score | 0.078888 (rank : 104) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
CALB1_HUMAN
|
||||||
| NC score | 0.078832 (rank : 105) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.078691 (rank : 106) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.078421 (rank : 107) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
GUC1B_HUMAN
|
||||||
| NC score | 0.077882 (rank : 108) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
KIP3_HUMAN
|
||||||
| NC score | 0.077101 (rank : 109) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96Q77 | Gene names | CIB3, KIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 3 (Kinase-interacting protein 3) (KIP 3). | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.076545 (rank : 110) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
EFCB3_MOUSE
|
||||||
| NC score | 0.076322 (rank : 111) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80X60 | Gene names | Efcab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
EFCB1_MOUSE
|
||||||
| NC score | 0.075784 (rank : 112) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.075605 (rank : 113) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.075605 (rank : 114) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
KIP1_HUMAN
|
||||||
| NC score | 0.074845 (rank : 115) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
KIP1_MOUSE
|
||||||
| NC score | 0.074379 (rank : 116) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.074045 (rank : 117) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.073409 (rank : 118) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.073409 (rank : 119) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
EFCB3_HUMAN
|
||||||
| NC score | 0.073340 (rank : 120) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N7B9 | Gene names | EFCAB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 3. | |||||
|
SORCN_HUMAN
|
||||||
| NC score | 0.071456 (rank : 121) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P30626 | Gene names | SRI | |||
|
Domain Architecture |

|
|||||
| Description | Sorcin (22 kDa protein) (CP-22) (V19). | |||||
|
HPCL4_HUMAN
|
||||||
| NC score | 0.071451 (rank : 122) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| NC score | 0.071449 (rank : 123) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.070380 (rank : 124) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
PLST_HUMAN
|
||||||
| NC score | 0.069491 (rank : 125) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13797, Q86YI6 | Gene names | PLS3 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
EFCB1_HUMAN
|
||||||
| NC score | 0.069412 (rank : 126) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
PLST_MOUSE
|
||||||
| NC score | 0.069012 (rank : 127) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99K51 | Gene names | Pls3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plastin-3 (T-plastin). | |||||
|
AIF1_HUMAN
|
||||||
| NC score | 0.067657 (rank : 128) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P55008, Q9UKS9 | Gene names | AIF1, G1, IBA1 | |||
|
Domain Architecture |

|
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1) (Protein G1). | |||||
|
AIF1_MOUSE
|
||||||
| NC score | 0.067191 (rank : 129) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70200 | Gene names | Aif1, Iba1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Allograft inflammatory factor 1 (AIF-1) (Ionized calcium-binding adapter molecule 1). | |||||
|
GUC1B_MOUSE
|
||||||
| NC score | 0.064950 (rank : 130) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
IBA2_MOUSE
|
||||||
| NC score | 0.063884 (rank : 131) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EQX4, Q8C150 | Gene names | Iba2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
RCN2_MOUSE
|
||||||
| NC score | 0.062966 (rank : 132) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BP92, O70341 | Gene names | Rcn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-2 precursor (Taipoxin-associated calcium-binding protein 49) (TCBP-49). | |||||
|
IBA2_HUMAN
|
||||||
| NC score | 0.062905 (rank : 133) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BQI0, Q6ZR40, Q8NAX7, Q8WU47, Q9H9G0 | Gene names | IBA2, C9orf58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ionized calcium-binding adapter molecule 2. | |||||
|
SORCN_MOUSE
|
||||||
| NC score | 0.062539 (rank : 134) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P069, Q3UKC5, Q9CR38, Q9D7V8 | Gene names | Sri | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorcin. | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.061272 (rank : 135) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
GRAN_HUMAN
|
||||||
| NC score | 0.060948 (rank : 136) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28676 | Gene names | GCA, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Grancalcin. | |||||
|
AYTL1_HUMAN
|
||||||
| NC score | 0.059861 (rank : 137) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
PEF1_HUMAN
|
||||||
| NC score | 0.059109 (rank : 138) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UBV8 | Gene names | PEF1, ABP32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.058788 (rank : 139) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.058788 (rank : 140) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
PPE2_MOUSE
|
||||||
| NC score | 0.058201 (rank : 141) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
MCFD2_HUMAN
|
||||||
| NC score | 0.057786 (rank : 142) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NI22, Q8N3M5 | Gene names | MCFD2, SDNSF | |||
|
Domain Architecture |

|
|||||
| Description | Multiple coagulation factor deficiency protein 2 precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
RCN2_HUMAN
|
||||||
| NC score | 0.057520 (rank : 143) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
AYT1A_MOUSE
|
||||||
| NC score | 0.057397 (rank : 144) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.055887 (rank : 145) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
RECO_MOUSE
|
||||||
| NC score | 0.055635 (rank : 146) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
KCIP4_HUMAN
|
||||||
| NC score | 0.055325 (rank : 147) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| NC score | 0.055311 (rank : 148) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
DUOX2_HUMAN
|
||||||
| NC score | 0.055269 (rank : 149) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
EFHC1_MOUSE
|
||||||
| NC score | 0.053701 (rank : 150) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9D9T8 | Gene names | Efhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.053495 (rank : 151) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.051869 (rank : 152) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
CPNS2_HUMAN
|
||||||
| NC score | 0.051751 (rank : 153) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96L46, Q9BPV4 | Gene names | CAPNS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain small subunit 2 (CSS2) (Calcium-dependent protease small subunit 2). | |||||
|
RECO_HUMAN
|
||||||
| NC score | 0.051464 (rank : 154) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
TNNT3_MOUSE
|
||||||
| NC score | 0.050430 (rank : 155) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
MCFD2_MOUSE
|
||||||
| NC score | 0.049297 (rank : 156) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K5B2, Q3U9G5 | Gene names | Mcfd2, Sdnsf | |||
|
Domain Architecture |

|
|||||
| Description | Multiple coagulation factor deficiency protein 2 homolog precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.044881 (rank : 157) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
NOC3L_HUMAN
|
||||||
| NC score | 0.042524 (rank : 158) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WTT2, Q9H5M6, Q9H9D8 | Gene names | NOC3L, AD24, C10orf117, FAD24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.038610 (rank : 159) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.037311 (rank : 160) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
RYR2_HUMAN
|
||||||
| NC score | 0.035376 (rank : 161) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92736, Q15411, Q546N8 | Gene names | RYR2 | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 2 (Cardiac muscle-type ryanodine receptor) (RyR2) (RYR-2) (Cardiac muscle ryanodine receptor-calcium release channel) (hRYR-2). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.034513 (rank : 162) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
PLCB3_HUMAN
|
||||||
| NC score | 0.032604 (rank : 163) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
NOC3L_MOUSE
|
||||||
| NC score | 0.031463 (rank : 164) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.030742 (rank : 165) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.030391 (rank : 166) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
LZTS1_HUMAN
|
||||||
| NC score | 0.030216 (rank : 167) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
SEL1L_HUMAN
|
||||||
| NC score | 0.029895 (rank : 168) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBV2, Q6UWT6, Q9P1T9, Q9UHK7 | Gene names | SEL1L, TSA305 | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
SEL1L_MOUSE
|
||||||
| NC score | 0.029759 (rank : 169) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2G6, Q9DBD8 | Gene names | Sel1l, Sel1h | |||
|
Domain Architecture |

|
|||||
| Description | Sel-1 homolog precursor (Suppressor of lin-12-like protein) (Sel-1L). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.029612 (rank : 170) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.028416 (rank : 171) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.026879 (rank : 172) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.022761 (rank : 173) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
ITIH4_HUMAN
|
||||||
| NC score | 0.022343 (rank : 174) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14624, Q15135, Q9P190, Q9UQ54 | Gene names | ITIH4, IHRP, ITIHL1, PK120 | |||
|
Domain Architecture |

|
|||||
| Description | Inter-alpha-trypsin inhibitor heavy chain H4 precursor (ITI heavy chain H4) (Inter-alpha-inhibitor heavy chain 4) (Inter-alpha-trypsin inhibitor family heavy chain-related protein) (IHRP) (Plasma kallikrein sensitive glycoprotein 120) (PK-120) (GP120) [Contains: 70 kDa inter-alpha-trypsin inhibitor heavy chain H4; 35 kDa inter-alpha- trypsin inhibitor heavy chain H4]. | |||||
|
MCES_MOUSE
|
||||||
| NC score | 0.021690 (rank : 175) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0L8, Q3V3U9, Q6ZQC6, Q9D5F1 | Gene names | Rnmt, Kiaa0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.020975 (rank : 176) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.020401 (rank : 177) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
KI21B_MOUSE
|
||||||
| NC score | 0.020274 (rank : 178) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXL1, P97424 | Gene names | Kif21b, Kif6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21B (Kinesin-like protein KIF6). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.020228 (rank : 179) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.017461 (rank : 180) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.009864 (rank : 181) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.007709 (rank : 182) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.005672 (rank : 183) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
ZBTB1_HUMAN
|
||||||
| NC score | 0.001528 (rank : 184) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2K1, Q86SW8 | Gene names | ZBTB1, KIAA0997 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||
|
ZBTB1_MOUSE
|
||||||
| NC score | 0.001076 (rank : 185) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91VL9, Q8CDP7, Q99LD2 | Gene names | Zbtb1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 1. | |||||