Please be patient as the page loads
|
MCFD2_HUMAN
|
||||||
| SwissProt Accessions | Q8NI22, Q8N3M5 | Gene names | MCFD2, SDNSF | |||
|
Domain Architecture |
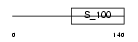
|
|||||
| Description | Multiple coagulation factor deficiency protein 2 precursor (Neural stem cell-derived neuronal survival protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCFD2_HUMAN
|
||||||
| θ value | 7.19216e-82 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NI22, Q8N3M5 | Gene names | MCFD2, SDNSF | |||
|
Domain Architecture |

|
|||||
| Description | Multiple coagulation factor deficiency protein 2 precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
MCFD2_MOUSE
|
||||||
| θ value | 4.68348e-65 (rank : 2) | NC score | 0.992367 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K5B2, Q3U9G5 | Gene names | Mcfd2, Sdnsf | |||
|
Domain Architecture |
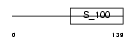
|
|||||
| Description | Multiple coagulation factor deficiency protein 2 homolog precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 3) | NC score | 0.339703 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 4) | NC score | 0.265637 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
KIP2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 5) | NC score | 0.339658 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 6) | NC score | 0.341850 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.229906 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
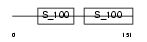
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
KIP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.321502 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |
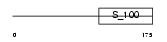
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
GUC1B_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 9) | NC score | 0.283561 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |
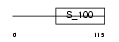
|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.299437 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |
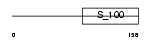
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.303542 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
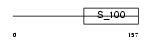
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.188634 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
CHP2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.242900 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |
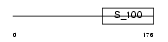
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.222909 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
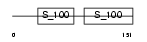
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
GUC1A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.294635 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |
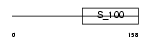
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
KIP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.302872 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |
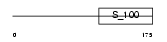
|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
NUCB2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.193856 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80303, Q8NFT5 | Gene names | NUCB2, NEFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA) (Gastric cancer antigen Zg4). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.268234 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
EFHB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.161587 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.214266 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.212849 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
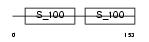
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
HPCL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.265829 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
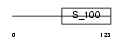
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.174706 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.176503 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.181917 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALM_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.185962 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.185962 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
KCIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.244391 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.243867 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
VISL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.250974 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.250974 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.154958 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
NUCB1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.148010 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
GUC1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.262228 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.258984 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.258984 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
HPCL4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.244801 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.244821 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.002215 (rank : 155) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.242930 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
DGKG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.100480 (rank : 95) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.175393 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
DGKB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.127756 (rank : 81) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6T7, O75241, Q75MF9, Q75MU7, Q86UI5, Q86UM9, Q9UQ29 | Gene names | DGKB, DAGK2, KIAA0718 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase beta (EC 2.7.1.107) (Diglyceride kinase beta) (DGK-beta) (DAG kinase beta) (90 kDa diacylglycerol kinase). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.025059 (rank : 152) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
NCALD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.252518 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |
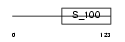
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.252518 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |
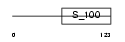
|
|||||
| Description | Neurocalcin delta. | |||||
|
CAYP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.057786 (rank : 145) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
CHP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.208441 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |
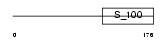
|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
DGKG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.092545 (rank : 102) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
RCN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.106676 (rank : 94) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BP92, O70341 | Gene names | Rcn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-2 precursor (Taipoxin-associated calcium-binding protein 49) (TCBP-49). | |||||
|
RCN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.082152 (rank : 112) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BH97, Q58E50, Q8R137, Q99K35, Q9CTD4 | Gene names | Rcn3, D7Ertd671e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-3 precursor. | |||||
|
CABP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.156801 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |
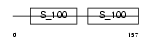
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CSEN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.243639 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CABP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.152301 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
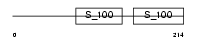
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
CABP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.143358 (rank : 66) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |
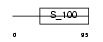
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.143358 (rank : 67) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |
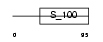
|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
KCIP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.233423 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.233856 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
PDIA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.018585 (rank : 153) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30101, Q13453, Q14255, Q8IYF8, Q9UMU7 | Gene names | PDIA3, ERP60, GRP58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (58 kDa glucose-regulated protein). | |||||
|
RCN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.077452 (rank : 117) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96D15, Q9HBZ8 | Gene names | RCN3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.027428 (rank : 151) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
PDIA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.017682 (rank : 154) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27773 | Gene names | Pdia3, Erp, Erp60, Grp58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57). | |||||
|
AYT1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.117158 (rank : 86) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
AYT1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.120842 (rank : 85) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.121036 (rank : 84) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
CABP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.134231 (rank : 77) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |
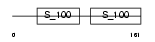
|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CABP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.134252 (rank : 76) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |
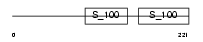
|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.152751 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |
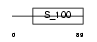
|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.134261 (rank : 75) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.137238 (rank : 73) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.139785 (rank : 70) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CABP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.140908 (rank : 68) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CALB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.131825 (rank : 79) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |
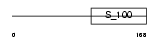
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CALB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.130783 (rank : 80) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |
|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
CALL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.114713 (rank : 88) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
CALM4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.160303 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CALN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.111166 (rank : 90) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.110775 (rank : 91) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |
|
|||||
| Description | Calneuron-1. | |||||
|
CALU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.068490 (rank : 129) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
CALU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.068252 (rank : 130) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.226712 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.226733 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.214193 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.224701 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
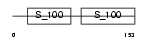
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CAYP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.139792 (rank : 69) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |
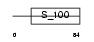
|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CETN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.139456 (rank : 71) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |
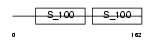
|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.137568 (rank : 72) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
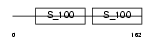
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.133576 (rank : 78) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.135224 (rank : 74) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.125862 (rank : 83) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.126484 (rank : 82) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
|
|||||
| Description | Centrin-3. | |||||
|
CHP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.182888 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99653 | Gene names | CHP | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog). | |||||
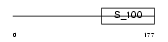
|
CHP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.185743 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
DGKA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.071570 (rank : 123) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23743, O75481, O75482, O75483 | Gene names | DGKA, DAGK, DAGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
DGKA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.095896 (rank : 99) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88673, Q922X2 | Gene names | Dgka, Dagk1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.116617 (rank : 87) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
DUOX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.108895 (rank : 92) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
EFCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.217897 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.216268 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.079648 (rank : 113) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFCB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.088596 (rank : 104) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
EFHB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.077932 (rank : 116) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
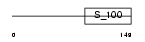
|
EFHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.075370 (rank : 120) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
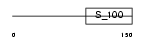
|
EFHD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.077192 (rank : 118) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
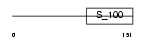
|
EFHD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.059160 (rank : 144) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96C19 | Gene names | EFHD2, SWS1 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
EFHD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.059529 (rank : 143) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D8Y0, Q9D0P4 | Gene names | Efhd2, Sws1 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.050508 (rank : 150) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.218990 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.216865 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.219586 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96Q77 | Gene names | CIB3, KIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 3 (Kinase-interacting protein 3) (KIP 3). | |||||
|
MLE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.066167 (rank : 135) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.065568 (rank : 137) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MLE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.066820 (rank : 131) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.066218 (rank : 134) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLR5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.072919 (rank : 122) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |
|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
MLRA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.070987 (rank : 125) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |
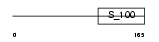
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.076342 (rank : 119) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |
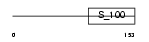
|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MLRM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.087488 (rank : 105) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |
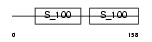
|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
MLRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.083651 (rank : 111) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |
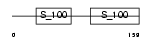
|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
MLRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.083850 (rank : 110) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.051447 (rank : 149) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |
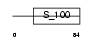
|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
MLRV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.062512 (rank : 140) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |
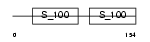
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.062269 (rank : 141) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |
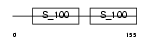
|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.066429 (rank : 133) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
MYL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.066802 (rank : 132) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MYL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.062016 (rank : 142) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
MYL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.063774 (rank : 138) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09541 | Gene names | Myl4, Mlc1a, Myla | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, atrial/fetal isoform) (MLC1A) (MLC1EMB). | |||||
|
MYL6B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.071508 (rank : 124) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
MYL6B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.068860 (rank : 128) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
MYL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.069710 (rank : 126) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
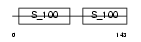
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.069710 (rank : 127) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
NCS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.231701 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.231701 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
NOX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.084394 (rank : 108) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
ONCO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.097737 (rank : 97) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
ONCO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.114141 (rank : 89) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |
|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
PCAT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.073272 (rank : 121) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
PCAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.086190 (rank : 106) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3TFD2, Q3TAX4, Q6NXZ6, Q8BG23, Q8BUX7, Q99JU6 | Gene names | Aytl2, Lpcat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (mLPCAT1) (Acyltransferase-like 2). | |||||
|
PDCD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.093543 (rank : 101) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
PDCD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.094111 (rank : 100) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |
|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.066133 (rank : 136) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
PLSL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.056628 (rank : 147) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
PLSL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.057360 (rank : 146) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PRVA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.090377 (rank : 103) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
PRVA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.097898 (rank : 96) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |
|
|||||
| Description | Parvalbumin alpha. | |||||
|
RCN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.062635 (rank : 139) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15293 | Gene names | RCN1, RCN | |||
|
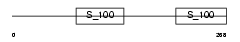
Domain Architecture |
|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
RCN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.054945 (rank : 148) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05186, Q3TVU3 | Gene names | Rcn1, Rca1, Rcn | |||
|
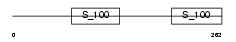
Domain Architecture |
|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
RCN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.083928 (rank : 109) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
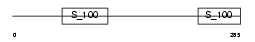
Domain Architecture |
|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
RECO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.206544 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
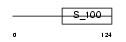
Domain Architecture |
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
RECO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.207102 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
SEGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.107731 (rank : 93) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
|
|||||
| Description | Secretagogin. | |||||
|
SEGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.097711 (rank : 98) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
TESC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.085094 (rank : 107) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96BS2 | Gene names | TESC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tescalcin (TSC). | |||||
|
TESC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.079242 (rank : 114) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKL5 | Gene names | Tesc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tescalcin (TSC). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.078175 (rank : 115) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
MCFD2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.19216e-82 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8NI22, Q8N3M5 | Gene names | MCFD2, SDNSF | |||
|
Domain Architecture |

|
|||||
| Description | Multiple coagulation factor deficiency protein 2 precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
MCFD2_MOUSE
|
||||||
| NC score | 0.992367 (rank : 2) | θ value | 4.68348e-65 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8K5B2, Q3U9G5 | Gene names | Mcfd2, Sdnsf | |||
|
Domain Architecture |

|
|||||
| Description | Multiple coagulation factor deficiency protein 2 homolog precursor (Neural stem cell-derived neuronal survival protein). | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.341850 (rank : 3) | θ value | 0.00035302 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.339703 (rank : 4) | θ value | 3.19293e-05 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
KIP2_HUMAN
|
||||||
| NC score | 0.339658 (rank : 5) | θ value | 0.000270298 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP1_MOUSE
|
||||||
| NC score | 0.321502 (rank : 6) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z0F4, Q3TN80 | Gene names | Cib1, Cib, Kip, Prkdcip | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB). | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.303542 (rank : 7) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
KIP1_HUMAN
|
||||||
| NC score | 0.302872 (rank : 8) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99828, O00693, O00735, Q6IB49, Q96J54, Q99971 | Gene names | CIB1, CIB, KIP, PRKDCIP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 1 (Calmyrin) (DNA-PKcs- interacting protein) (Kinase-interacting protein) (KIP) (CIB) (SNK- interacting protein 2-28) (SIP2-28). | |||||
|
GUC1A_HUMAN
|
||||||
| NC score | 0.299437 (rank : 9) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1A_MOUSE
|
||||||
| NC score | 0.294635 (rank : 10) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1B_HUMAN
|
||||||
| NC score | 0.283561 (rank : 11) | θ value | 0.0193708 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.268234 (rank : 12) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.265829 (rank : 13) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.265637 (rank : 14) | θ value | 0.000121331 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
GUC1B_MOUSE
|
||||||
| NC score | 0.262228 (rank : 15) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.258984 (rank : 16) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.258984 (rank : 17) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
NCALD_HUMAN
|
||||||
| NC score | 0.252518 (rank : 18) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| NC score | 0.252518 (rank : 19) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.250974 (rank : 20) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.250974 (rank : 21) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
HPCL4_MOUSE
|
||||||
| NC score | 0.244821 (rank : 22) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
HPCL4_HUMAN
|
||||||
| NC score | 0.244801 (rank : 23) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
KCIP1_HUMAN
|
||||||
| NC score | 0.244391 (rank : 24) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| NC score | 0.243867 (rank : 25) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.243639 (rank : 26) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.242930 (rank : 27) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
CHP2_HUMAN
|
||||||
| NC score | 0.242900 (rank : 28) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O43745 | Gene names | CHP2, HCA520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520). | |||||
|
KCIP4_MOUSE
|
||||||
| NC score | 0.233856 (rank : 29) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_HUMAN
|
||||||
| NC score | 0.233423 (rank : 30) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.231701 (rank : 31) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.231701 (rank : 32) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.229906 (rank : 33) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.226733 (rank : 34) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.226712 (rank : 35) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.224701 (rank : 36) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.222909 (rank : 37) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
KIP3_HUMAN
|
||||||
| NC score | 0.219586 (rank : 38) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96Q77 | Gene names | CIB3, KIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium and integrin-binding protein 3 (Kinase-interacting protein 3) (KIP 3). | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.218990 (rank : 39) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
EFCB1_HUMAN
|
||||||
| NC score | 0.217897 (rank : 40) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.216865 (rank : 41) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
EFCB1_MOUSE
|
||||||
| NC score | 0.216268 (rank : 42) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.214266 (rank : 43) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.214193 (rank : 44) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.212849 (rank : 45) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
CHP2_MOUSE
|
||||||
| NC score | 0.208441 (rank : 46) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D869 | Gene names | Chp2, Hca520 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin B homologous protein 2 (Hepatocellular carcinoma- associated antigen 520 homolog). | |||||
|
RECO_MOUSE
|
||||||
| NC score | 0.207102 (rank : 47) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
RECO_HUMAN
|
||||||
| NC score | 0.206544 (rank : 48) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
NUCB2_HUMAN
|
||||||
| NC score | 0.193856 (rank : 49) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80303, Q8NFT5 | Gene names | NUCB2, NEFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA) (Gastric cancer antigen Zg4). | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.188634 (rank : 50) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.185962 (rank : 51) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.185962 (rank : 52) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CHP1_MOUSE
|
||||||
| NC score | 0.185743 (rank : 53) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P61022, Q62877, Q6ZWQ8 | Gene names | Chp, Sid470 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Sid 470). | |||||
|
CHP1_HUMAN
|
||||||
| NC score | 0.182888 (rank : 54) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99653 | Gene names | CHP | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein p22 (Calcium-binding protein CHP) (Calcineurin homologous protein) (Calcineurin B homolog). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.181917 (rank : 55) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.176503 (rank : 56) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.175393 (rank : 57) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.174706 (rank : 58) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
EFHB_HUMAN
|
||||||
| NC score | 0.161587 (rank : 59) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N7U6, Q6ZWK9, Q8IV58, Q96LQ6 | Gene names | EFHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CALM4_MOUSE
|
||||||
| NC score | 0.160303 (rank : 60) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JM83, Q9CR31, Q9D1E9 | Gene names | Calm4, Dd112 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-4 (Calcium-binding protein Dd112). | |||||
|
CABP2_MOUSE
|
||||||
| NC score | 0.156801 (rank : 61) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLK4, Q9JLK5 | Gene names | Cabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.154958 (rank : 62) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
CABP3_HUMAN
|
||||||
| NC score | 0.152751 (rank : 63) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZU6, Q9NYY0, Q9NZT9 | Gene names | CABP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 3 (CaBP3). | |||||
|
CABP2_HUMAN
|
||||||
| NC score | 0.152301 (rank : 64) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NPB3 | Gene names | CABP2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcium-binding protein 2 (CaBP2). | |||||
|
NUCB1_MOUSE
|
||||||
| NC score | 0.148010 (rank : 65) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02819, Q3TU42, Q9CWA1, Q9D8J4, Q9DBL3 | Gene names | Nucb1, Nuc, Nucb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
CABP7_HUMAN
|
||||||
| NC score | 0.143358 (rank : 66) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q86V35 | Gene names | CABP7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP7_MOUSE
|
||||||
| NC score | 0.143358 (rank : 67) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q91ZM8 | Gene names | Cabp7 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 7 (CaBP7). | |||||
|
CABP5_MOUSE
|
||||||
| NC score | 0.140908 (rank : 68) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JLK3 | Gene names | Cabp5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CAYP1_HUMAN
|
||||||
| NC score | 0.139792 (rank : 69) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13938, Q3MJA7, Q8WUZ3 | Gene names | CAPS | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin (Calcyphosine). | |||||
|
CABP5_HUMAN
|
||||||
| NC score | 0.139785 (rank : 70) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NP86 | Gene names | CABP5 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 5 (CaBP5). | |||||
|
CETN1_HUMAN
|
||||||
| NC score | 0.139456 (rank : 71) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12798 | Gene names | CETN1, CEN1, CETN | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin isoform 2). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.137568 (rank : 72) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CABP4_MOUSE
|
||||||
| NC score | 0.137238 (rank : 73) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VHC5 | Gene names | Cabp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.135224 (rank : 74) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.134261 (rank : 75) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CABP1_MOUSE
|
||||||
| NC score | 0.134252 (rank : 76) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLK7, Q9JLK6 | Gene names | Cabp1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1). | |||||
|
CABP1_HUMAN
|
||||||
| NC score | 0.134231 (rank : 77) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZU7, O95663, Q9NZU8 | Gene names | CABP1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 1 (CaBP1) (Calbrain). | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.133576 (rank : 78) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
CALB1_HUMAN
|
||||||
| NC score | 0.131825 (rank : 79) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P05937 | Gene names | CALB1, CAB27 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K). | |||||
|
CALB1_MOUSE
|
||||||
| NC score | 0.130783 (rank : 80) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12658, Q545N6 | Gene names | Calb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calbindin (Vitamin D-dependent calcium-binding protein, avian-type) (Calbindin D28) (D-28K) (Spot 35 protein) (PCD-29). | |||||
|
DGKB_HUMAN
|
||||||
| NC score | 0.127756 (rank : 81) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y6T7, O75241, Q75MF9, Q75MU7, Q86UI5, Q86UM9, Q9UQ29 | Gene names | DGKB, DAGK2, KIAA0718 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase beta (EC 2.7.1.107) (Diglyceride kinase beta) (DGK-beta) (DAG kinase beta) (90 kDa diacylglycerol kinase). | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.126484 (rank : 82) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.125862 (rank : 83) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
AYTL1_HUMAN
|
||||||
| NC score | 0.121036 (rank : 84) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
AYT1B_MOUSE
|
||||||
| NC score | 0.120842 (rank : 85) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
AYT1A_MOUSE
|
||||||
| NC score | 0.117158 (rank : 86) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.116617 (rank : 87) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
CALL6_HUMAN
|
||||||
| NC score | 0.114713 (rank : 88) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TD86, Q6Q2C4 | Gene names | CALML6, CAGLP, CALGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-like protein 6 (Calglandulin-like protein). | |||||
|
ONCO_MOUSE
|
||||||
| NC score | 0.114141 (rank : 89) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51879, Q62004 | Gene names | Ocm | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
CALN1_HUMAN
|
||||||
| NC score | 0.111166 (rank : 90) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BXU9 | Gene names | CALN1, CABP8 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1 (Calcium-binding protein CaBP8). | |||||
|
CALN1_MOUSE
|
||||||
| NC score | 0.110775 (rank : 91) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JJG7 | Gene names | Caln1 | |||
|
Domain Architecture |

|
|||||
| Description | Calneuron-1. | |||||
|
DUOX2_HUMAN
|
||||||
| NC score | 0.108895 (rank : 92) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.107731 (rank : 93) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
RCN2_MOUSE
|
||||||
| NC score | 0.106676 (rank : 94) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BP92, O70341 | Gene names | Rcn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-2 precursor (Taipoxin-associated calcium-binding protein 49) (TCBP-49). | |||||
|
DGKG_MOUSE
|
||||||
| NC score | 0.100480 (rank : 95) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91WG7 | Gene names | Dgkg, Dagk3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma) (88 kDa diacylglycerol kinase). | |||||
|
PRVA_MOUSE
|
||||||
| NC score | 0.097898 (rank : 96) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P32848 | Gene names | Pvalb, Pva | |||
|
Domain Architecture |

|
|||||
| Description | Parvalbumin alpha. | |||||
|
ONCO_HUMAN
|
||||||
| NC score | 0.097737 (rank : 97) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P32930 | Gene names | OCM, OCMN | |||
|
Domain Architecture |

|
|||||
| Description | Oncomodulin (OM) (Parvalbumin beta). | |||||
|
SEGN_MOUSE
|
||||||
| NC score | 0.097711 (rank : 98) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
DGKA_MOUSE
|
||||||
| NC score | 0.095896 (rank : 99) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88673, Q922X2 | Gene names | Dgka, Dagk1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
PDCD6_MOUSE
|
||||||
| NC score | 0.094111 (rank : 100) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12815, Q545I0, Q61145 | Gene names | Pdcd6, Alg2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2) (PMP41) (ALG-257). | |||||
|
PDCD6_HUMAN
|
||||||
| NC score | 0.093543 (rank : 101) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75340 | Gene names | PDCD6, ALG2 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 6 (Probable calcium-binding protein ALG- 2). | |||||
|
DGKG_HUMAN
|
||||||
| NC score | 0.092545 (rank : 102) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P49619 | Gene names | DGKG, DAGK3 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase gamma (EC 2.7.1.107) (Diglyceride kinase gamma) (DGK-gamma) (DAG kinase gamma). | |||||
|
PRVA_HUMAN
|
||||||
| NC score | 0.090377 (rank : 103) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20472, P78378, Q4VB78, Q5R3Q9 | Gene names | PVALB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parvalbumin alpha. | |||||
|
EFCB2_MOUSE
|
||||||
| NC score | 0.088596 (rank : 104) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQ46, Q9CS07 | Gene names | Efcab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
MLRM_HUMAN
|
||||||
| NC score | 0.087488 (rank : 105) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P19105 | Gene names | MRLC2, MLCB, RLC | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, nonsarcomeric (Myosin RLC). | |||||
|
PCAT1_MOUSE
|
||||||
| NC score | 0.086190 (rank : 106) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3TFD2, Q3TAX4, Q6NXZ6, Q8BG23, Q8BUX7, Q99JU6 | Gene names | Aytl2, Lpcat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (mLPCAT1) (Acyltransferase-like 2). | |||||
|
TESC_HUMAN
|
||||||
| NC score | 0.085094 (rank : 107) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96BS2 | Gene names | TESC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tescalcin (TSC). | |||||
|
NOX5_HUMAN
|
||||||
| NC score | 0.084394 (rank : 108) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
RCN2_HUMAN
|
||||||
| NC score | 0.083928 (rank : 109) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14257 | Gene names | RCN2, ERC55 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-2 precursor (Calcium-binding protein ERC-55) (E6- binding protein) (E6BP). | |||||
|
MLRN_MOUSE
|
||||||
| NC score | 0.083850 (rank : 110) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9CQ19, Q80X77 | Gene names | Myl9, Myrl2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9). | |||||
|
MLRN_HUMAN
|
||||||
| NC score | 0.083651 (rank : 111) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24844, Q9H136 | Gene names | MYL9, MLC2, MRLC1, MYRL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, smooth muscle isoform (Myosin RLC) (Myosin regulatory light chain 9) (LC20). | |||||
|
RCN3_MOUSE
|
||||||
| NC score | 0.082152 (rank : 112) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BH97, Q58E50, Q8R137, Q99K35, Q9CTD4 | Gene names | Rcn3, D7Ertd671e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulocalbin-3 precursor. | |||||
|
EFCB2_HUMAN
|
||||||
| NC score | 0.079648 (rank : 113) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VUJ9, Q59G23, Q9BS36 | Gene names | EFCAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 2. | |||||
|
TESC_MOUSE
|
||||||
| NC score | 0.079242 (rank : 114) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKL5 | Gene names | Tesc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tescalcin (TSC). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.078175 (rank : 115) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.077932 (rank : 116) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
RCN3_HUMAN
|
||||||
| NC score | 0.077452 (rank : 117) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96D15, Q9HBZ8 | Gene names | RCN3 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-3 precursor (EF-hand calcium-binding protein RLP49). | |||||
|
EFHD1_MOUSE
|
||||||
| NC score | 0.077192 (rank : 118) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D4J1, Q543U4 | Gene names | Efhd1, Sws2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
MLRA_MOUSE
|
||||||
| NC score | 0.076342 (rank : 119) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QVP4, Q63977, Q9JIE8 | Gene names | Myl7, Mylc2a | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
EFHD1_HUMAN
|
||||||
| NC score | 0.075370 (rank : 120) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
PCAT1_HUMAN
|
||||||
| NC score | 0.073272 (rank : 121) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
MLR5_HUMAN
|
||||||
| NC score | 0.072919 (rank : 122) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02045 | Gene names | MYL5 | |||
|
Domain Architecture |

|
|||||
| Description | Superfast myosin regulatory light chain 2 (MyLC-2) (MYLC2) (Myosin regulatory light chain 5). | |||||
|
DGKA_HUMAN
|
||||||
| NC score | 0.071570 (rank : 123) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23743, O75481, O75482, O75483 | Gene names | DGKA, DAGK, DAGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol kinase alpha (EC 2.7.1.107) (Diglyceride kinase alpha) (DGK-alpha) (DAG kinase alpha) (80 kDa diacylglycerol kinase). | |||||
|
MYL6B_HUMAN
|
||||||
| NC score | 0.071508 (rank : 124) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14649 | Gene names | MYL6B, MLC1SA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B) (Myosin light chain 1 slow-twitch muscle A isoform) (MLC1sa). | |||||
|
MLRA_HUMAN
|
||||||
| NC score | 0.070987 (rank : 125) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01449 | Gene names | MYL7, MYLC2A | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, atrial isoform (Myosin light chain 2a) (MLC-2a) (MLC2a) (Myosin regulatory light chain 7). | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.069710 (rank : 126) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.069710 (rank : 127) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6B_MOUSE
|
||||||
| NC score | 0.068860 (rank : 128) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CI43 | Gene names | Myl6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light polypeptide 6B (Smooth muscle and nonmuscle myosin light chain alkali 6B). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.068490 (rank : 129) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
CALU_MOUSE
|
||||||
| NC score | 0.068252 (rank : 130) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
MLE3_HUMAN
|
||||||
| NC score | 0.066820 (rank : 131) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P06741 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MYL3_MOUSE
|
||||||
| NC score | 0.066802 (rank : 132) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09542, Q3UIF4, Q9CQZ2 | Gene names | Myl3, Mlc1v, Mylc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain). | |||||
|
MYL3_HUMAN
|
||||||
| NC score | 0.066429 (rank : 133) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P08590, Q9NRS8 | Gene names | MYL3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 3 (Myosin light chain 1, slow-twitch muscle B/ventricular isoform) (MLC1SB) (Ventricular/slow twitch myosin alkali light chain) (Cardiac myosin light chain-1) (CMLC1). | |||||
|
MLE3_MOUSE
|
||||||
| NC score | 0.066218 (rank : 134) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05978 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 3, skeletal muscle isoform (A2 catalytic) (Alkali myosin light chain 3) (MLC3F). | |||||
|
MLE1_HUMAN
|
||||||
| NC score | 0.066167 (rank : 135) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05976 | Gene names | MYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.066133 (rank : 136) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
MLE1_MOUSE
|
||||||
| NC score | 0.065568 (rank : 137) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MYL4_MOUSE
|
||||||
| NC score | 0.063774 (rank : 138) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09541 | Gene names | Myl4, Mlc1a, Myla | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, atrial/fetal isoform) (MLC1A) (MLC1EMB). | |||||
|
RCN1_HUMAN
|
||||||
| NC score | 0.062635 (rank : 139) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15293 | Gene names | RCN1, RCN | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
MLRV_HUMAN
|
||||||
| NC score | 0.062512 (rank : 140) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10916, Q16123 | Gene names | MYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MLRV_MOUSE
|
||||||
| NC score | 0.062269 (rank : 141) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51667 | Gene names | Myl2, Mylpc | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2) (MLC-2v). | |||||
|
MYL4_HUMAN
|
||||||
| NC score | 0.062016 (rank : 142) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
EFHD2_MOUSE
|
||||||
| NC score | 0.059529 (rank : 143) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D8Y0, Q9D0P4 | Gene names | Efhd2, Sws1 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
EFHD2_HUMAN
|
||||||
| NC score | 0.059160 (rank : 144) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96C19 | Gene names | EFHD2, SWS1 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 2 (Swiprosin-1). | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.057786 (rank : 145) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
PLSL_MOUSE
|
||||||
| NC score | 0.057360 (rank : 146) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61233, Q3UE24, Q8R1X3, Q9CV77 | Gene names | Lcp1, Pls2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (65 kDa macrophage protein) (pp65). | |||||
|
PLSL_HUMAN
|
||||||
| NC score | 0.056628 (rank : 147) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13796 | Gene names | LCP1, PLS2 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-2 (L-plastin) (Lymphocyte cytosolic protein 1) (LCP-1) (LC64P). | |||||
|
RCN1_MOUSE
|
||||||
| NC score | 0.054945 (rank : 148) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05186, Q3TVU3 | Gene names | Rcn1, Rca1, Rcn | |||
|
Domain Architecture |

|
|||||
| Description | Reticulocalbin-1 precursor. | |||||
|
MLRS_MOUSE
|
||||||
| NC score | 0.051447 (rank : 149) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97457 | Gene names | Mylpf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin regulatory light chain 2, skeletal muscle isoform (MLC2F). | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.050508 (rank : 150) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.027428 (rank : 151) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.025059 (rank : 152) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
PDIA3_HUMAN
|
||||||
| NC score | 0.018585 (rank : 153) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30101, Q13453, Q14255, Q8IYF8, Q9UMU7 | Gene names | PDIA3, ERP60, GRP58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (58 kDa glucose-regulated protein). | |||||
|
PDIA3_MOUSE
|
||||||
| NC score | 0.017682 (rank : 154) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27773 | Gene names | Pdia3, Erp, Erp60, Grp58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.002215 (rank : 155) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||