Please be patient as the page loads
|
DHX36_MOUSE
|
||||||
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX36_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994097 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 6.54689e-176 (rank : 3) | NC score | 0.960686 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 1.15563e-172 (rank : 4) | NC score | 0.962172 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 1.6163e-166 (rank : 5) | NC score | 0.959894 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 8.02161e-166 (rank : 6) | NC score | 0.962081 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 5.9766e-145 (rank : 7) | NC score | 0.965279 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 3.62588e-142 (rank : 8) | NC score | 0.962982 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 1.09172e-138 (rank : 9) | NC score | 0.957159 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 2.97304e-136 (rank : 10) | NC score | 0.956653 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 2.54619e-95 (rank : 11) | NC score | 0.927727 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 7.93343e-89 (rank : 12) | NC score | 0.908000 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 5.32145e-85 (rank : 13) | NC score | 0.921505 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 5.32145e-85 (rank : 14) | NC score | 0.921333 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 6.30065e-78 (rank : 15) | NC score | 0.920299 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 6.52014e-75 (rank : 16) | NC score | 0.910523 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 1.45253e-74 (rank : 17) | NC score | 0.911173 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 7.47731e-63 (rank : 18) | NC score | 0.906958 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 2.65945e-60 (rank : 19) | NC score | 0.903714 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 7.25919e-50 (rank : 20) | NC score | 0.870657 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 21) | NC score | 0.853913 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
DHX34_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 22) | NC score | 0.926410 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 2.76489e-41 (rank : 23) | NC score | 0.902339 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 24) | NC score | 0.896492 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
TTF1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.041637 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
SK2L2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 26) | NC score | 0.051944 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 27) | NC score | 0.042223 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
DDX47_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.030535 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.031185 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.041497 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.042766 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.037011 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SK2L2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.049904 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.020683 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.024854 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.046750 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.058528 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.016311 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.016115 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.018909 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
DDX49_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.036942 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.012963 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.033472 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
SMBP2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.054623 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
FOLC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.021883 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48760 | Gene names | Fpgs | |||
|
Domain Architecture |
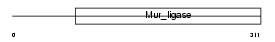
|
|||||
| Description | Folylpolyglutamate synthase, mitochondrial precursor (EC 6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS) (Tetrahydrofolate synthase) (Tetrahydrofolylpolyglutamate synthase). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.020621 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.022742 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.022042 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.025777 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.62314 (rank : 50) | NC score | 0.007328 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.020061 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.022640 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.012028 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.029738 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
REXO4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.017803 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PAQ4, Q3UU17, Q8C524 | Gene names | Rexo4, Gm111, Pmc2, Xpmc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.016186 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
ZBT11_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | -0.002936 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95625 | Gene names | ZBTB11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 11. | |||||
|
ASPM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.018360 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.013111 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.016689 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.014773 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.015819 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.027957 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.005820 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
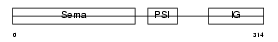
|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.010413 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.015830 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.012456 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
MACOI_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.022604 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MACOI_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.022297 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
LAMC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.005103 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
TRAP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.013019 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
DDX31_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.025776 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.043357 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DYXC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.014161 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.021381 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.015669 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.014596 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.011995 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
SEM3D_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.004861 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.016356 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.030842 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.010375 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.031655 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
PEX26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.016653 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z412, Q7Z413, Q7Z414, Q7Z415, Q7Z416, Q96B12, Q9NWQ0, Q9NXU0 | Gene names | PEX26 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 26 (Peroxin-26). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.003337 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.006592 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.006206 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.005912 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.005854 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.003925 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.016239 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FGD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.003383 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
G45IP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.014712 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TAE8, Q8IVM3, Q8TE51, Q969P9, Q9BSM6 | Gene names | GADD45GIP1, PLINP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 (CR6-interacting factor 1) (CRIF1) (CKII beta-associating protein) (Papillomavirus L2-interacting nuclear protein 1) (PLINP-1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.013668 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.007460 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.014230 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.022538 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAYP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.005672 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.024683 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.025093 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
E41L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.002687 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | -0.001536 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FSBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.007075 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKE5, Q8BJX8 | Gene names | Fsbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen silencer-binding protein. | |||||
|
G45IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.012224 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR59, Q8BT05 | Gene names | Gadd45gip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1. | |||||
|
K0776_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.006043 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94874, Q8N765, Q9NTQ0 | Gene names | KIAA0776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0776. | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.008461 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.006762 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
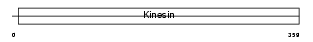
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.015790 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007454 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.026052 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.015518 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.011925 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.007367 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
TRAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.008054 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQN1, Q542I4 | Gene names | Trap1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.994097 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.965279 (rank : 3) | θ value | 5.9766e-145 (rank : 7) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.962982 (rank : 4) | θ value | 3.62588e-142 (rank : 8) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.962172 (rank : 5) | θ value | 1.15563e-172 (rank : 4) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.962081 (rank : 6) | θ value | 8.02161e-166 (rank : 6) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.960686 (rank : 7) | θ value | 6.54689e-176 (rank : 3) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.959894 (rank : 8) | θ value | 1.6163e-166 (rank : 5) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.957159 (rank : 9) | θ value | 1.09172e-138 (rank : 9) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.956653 (rank : 10) | θ value | 2.97304e-136 (rank : 10) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.927727 (rank : 11) | θ value | 2.54619e-95 (rank : 11) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.926410 (rank : 12) | θ value | 1.73182e-43 (rank : 22) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.921505 (rank : 13) | θ value | 5.32145e-85 (rank : 13) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.921333 (rank : 14) | θ value | 5.32145e-85 (rank : 14) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.920299 (rank : 15) | θ value | 6.30065e-78 (rank : 15) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.911173 (rank : 16) | θ value | 1.45253e-74 (rank : 17) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.910523 (rank : 17) | θ value | 6.52014e-75 (rank : 16) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.908000 (rank : 18) | θ value | 7.93343e-89 (rank : 12) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.906958 (rank : 19) | θ value | 7.47731e-63 (rank : 18) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.903714 (rank : 20) | θ value | 2.65945e-60 (rank : 19) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.902339 (rank : 21) | θ value | 2.76489e-41 (rank : 23) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.896492 (rank : 22) | θ value | 4.71619e-41 (rank : 24) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.870657 (rank : 23) | θ value | 7.25919e-50 (rank : 20) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.853913 (rank : 24) | θ value | 2.3352e-48 (rank : 21) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.058528 (rank : 25) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
SMBP2_MOUSE
|
||||||
| NC score | 0.054623 (rank : 26) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P40694 | Gene names | Ighmbp2, Smbp-2, Smbp2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Cardiac transcription factor 1) (CATF1). | |||||
|
SK2L2_MOUSE
|
||||||
| NC score | 0.051944 (rank : 27) | θ value | 0.0252991 (rank : 26) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZU3 | Gene names | Skiv2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
SK2L2_HUMAN
|
||||||
| NC score | 0.049904 (rank : 28) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42285, Q8TAG2 | Gene names | SKIV2L2, KIAA0052 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Superkiller viralicidic activity 2-like 2 (EC 3.6.1.-) (ATP-dependent helicase SKIV2L2). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.046750 (rank : 29) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.043357 (rank : 30) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.042766 (rank : 31) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.042223 (rank : 32) | θ value | 0.0330416 (rank : 27) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TTF1_HUMAN
|
||||||
| NC score | 0.041637 (rank : 33) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15361, Q4VXF3, Q58EY2, Q6P5T5 | Gene names | TTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (RNA polymerase I termination factor). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.041497 (rank : 34) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.037011 (rank : 35) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
DDX49_MOUSE
|
||||||
| NC score | 0.036942 (rank : 36) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q4FZF3 | Gene names | Ddx49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.033472 (rank : 37) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
PEO1_MOUSE
|
||||||
| NC score | 0.031655 (rank : 38) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.031185 (rank : 39) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.030842 (rank : 40) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX47_HUMAN
|
||||||
| NC score | 0.030535 (rank : 41) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0S4, Q96GM0, Q96NV8 | Gene names | DDX47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.029738 (rank : 42) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.027957 (rank : 43) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
PEO1_HUMAN
|
||||||
| NC score | 0.026052 (rank : 44) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.025777 (rank : 45) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
DDX31_HUMAN
|
||||||
| NC score | 0.025776 (rank : 46) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H8H2, Q5K6N2, Q5K6N3, Q5K6N4, Q5VZJ4, Q5VZJ9, Q96E91, Q96NY2, Q96SX5, Q9H5K6 | Gene names | DDX31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX31 (EC 3.6.1.-) (DEAD box protein 31) (Helicain). | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.025093 (rank : 47) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.024854 (rank : 48) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.024683 (rank : 49) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.022742 (rank : 50) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.022640 (rank : 51) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.022604 (rank : 52) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.022538 (rank : 53) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
MACOI_MOUSE
|
||||||
| NC score | 0.022297 (rank : 54) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.022042 (rank : 55) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
FOLC_MOUSE
|
||||||
| NC score | 0.021883 (rank : 56) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48760 | Gene names | Fpgs | |||
|
Domain Architecture |

|
|||||
| Description | Folylpolyglutamate synthase, mitochondrial precursor (EC 6.3.2.17) (Folylpoly-gamma-glutamate synthetase) (FPGS) (Tetrahydrofolate synthase) (Tetrahydrofolylpolyglutamate synthase). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.021381 (rank : 57) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.020683 (rank : 58) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.020621 (rank : 59) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.020061 (rank : 60) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.018909 (rank : 61) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
ASPM_HUMAN
|
||||||
| NC score | 0.018360 (rank : 62) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZT6, Q86UX4, Q8IUL2, Q8IZJ7, Q8IZJ8, Q8IZJ9, Q8N4D1, Q9NVS1, Q9NVT6 | Gene names | ASPM, MCPH5 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein (Abnormal spindle protein homolog) (Asp homolog). | |||||
|
REXO4_MOUSE
|
||||||
| NC score | 0.017803 (rank : 63) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PAQ4, Q3UU17, Q8C524 | Gene names | Rexo4, Gm111, Pmc2, Xpmc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.016689 (rank : 64) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PEX26_HUMAN
|
||||||
| NC score | 0.016653 (rank : 65) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z412, Q7Z413, Q7Z414, Q7Z415, Q7Z416, Q96B12, Q9NWQ0, Q9NXU0 | Gene names | PEX26 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 26 (Peroxin-26). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.016356 (rank : 66) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.016311 (rank : 67) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.016239 (rank : 68) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.016186 (rank : 69) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.016115 (rank : 70) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.015830 (rank : 71) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.015819 (rank : 72) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.015790 (rank : 73) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.015669 (rank : 74) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.015518 (rank : 75) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.014773 (rank : 76) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
G45IP_HUMAN
|
||||||
| NC score | 0.014712 (rank : 77) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TAE8, Q8IVM3, Q8TE51, Q969P9, Q9BSM6 | Gene names | GADD45GIP1, PLINP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 (CR6-interacting factor 1) (CRIF1) (CKII beta-associating protein) (Papillomavirus L2-interacting nuclear protein 1) (PLINP-1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.014596 (rank : 78) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.014230 (rank : 79) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
DYXC1_HUMAN
|
||||||
| NC score | 0.014161 (rank : 80) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WXU2, Q6P5Y9 | Gene names | DYX1C1, EKN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dyslexia susceptibility 1 candidate gene 1 protein. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.013668 (rank : 81) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.013111 (rank : 82) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
TRAP1_HUMAN
|
||||||
| NC score | 0.013019 (rank : 83) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.012963 (rank : 84) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.012456 (rank : 85) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
G45IP_MOUSE
|
||||||
| NC score | 0.012224 (rank : 86) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR59, Q8BT05 | Gene names | Gadd45gip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth arrest and DNA-damage-inducible proteins-interacting protein 1. | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.012028 (rank : 87) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.011995 (rank : 88) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.011925 (rank : 89) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.010413 (rank : 90) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.010375 (rank : 91) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.008461 (rank : 92) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
TRAP1_MOUSE
|
||||||
| NC score | 0.008054 (rank : 93) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQN1, Q542I4 | Gene names | Trap1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.007460 (rank : 94) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.007454 (rank : 95) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.007367 (rank : 96) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.007328 (rank : 97) | θ value | 0.62314 (rank : 50) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
FSBP_MOUSE
|
||||||
| NC score | 0.007075 (rank : 98) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BKE5, Q8BJX8 | Gene names | Fsbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen silencer-binding protein. | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.006762 (rank : 99) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.006592 (rank : 100) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.006206 (rank : 101) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
K0776_HUMAN
|
||||||
| NC score | 0.006043 (rank : 102) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94874, Q8N765, Q9NTQ0 | Gene names | KIAA0776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0776. | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.005912 (rank : 103) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.005854 (rank : 104) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.005820 (rank : 105) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
CAYP2_HUMAN
|
||||||
| NC score | 0.005672 (rank : 106) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXY5, Q6PH84, Q8N242, Q8NAY5 | Gene names | CAPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcyphosin-2 (Calcyphosine-2). | |||||
|
LAMC1_MOUSE
|
||||||
| NC score | 0.005103 (rank : 107) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
SEM3D_MOUSE
|
||||||
| NC score | 0.004861 (rank : 108) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.003925 (rank : 109) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
FGD3_HUMAN
|
||||||
| NC score | 0.003383 (rank : 110) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5JSP0, Q7Z7D9, Q8N5G1 | Gene names | FGD3, ZFYVE5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3 (Zinc finger FYVE domain-containing protein 5). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.003337 (rank : 111) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
E41L1_HUMAN
|
||||||
| NC score | 0.002687 (rank : 112) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4G0, O15046, Q4VXN4 | Gene names | EPB41L1, KIAA0338 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 1 (Neuronal protein 4.1) (4.1N). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | -0.001536 (rank : 113) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
ZBT11_HUMAN
|
||||||
| NC score | -0.002936 (rank : 114) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 114 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95625 | Gene names | ZBTB11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 11. | |||||