Please be patient as the page loads
|
DHX34_HUMAN
|
||||||
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX34_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 1.55827e-177 (rank : 2) | NC score | 0.973099 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 3) | NC score | 0.909873 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 3.15612e-45 (rank : 4) | NC score | 0.893274 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 3.15612e-45 (rank : 5) | NC score | 0.907324 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 1.73182e-43 (rank : 6) | NC score | 0.926410 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 2.95404e-43 (rank : 7) | NC score | 0.909736 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 8) | NC score | 0.930482 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 9) | NC score | 0.910141 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 10) | NC score | 0.926465 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 11) | NC score | 0.932859 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 12) | NC score | 0.932689 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 13) | NC score | 0.923518 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 4.41421e-39 (rank : 14) | NC score | 0.930417 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 15) | NC score | 0.925799 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 16) | NC score | 0.928370 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 17) | NC score | 0.924862 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 18) | NC score | 0.921950 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 19) | NC score | 0.934407 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 4.13158e-37 (rank : 20) | NC score | 0.920831 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 2.05049e-36 (rank : 21) | NC score | 0.919950 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 22) | NC score | 0.920345 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 23) | NC score | 0.912214 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 24) | NC score | 0.774610 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
PNKP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.051368 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLV6, Q6PFA3 | Gene names | Pnkp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
PNKP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.039242 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96T60, Q9P1V2, Q9UKU8, Q9UNF8 | Gene names | PNKP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.014475 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
DSG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.003552 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SJ6, Q6Y9L9, Q8IXV4 | Gene names | DSG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
HPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.015623 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92902, O15402, O15502 | Gene names | HPS1, HPS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein. | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.004317 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.008050 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.022697 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
KAD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.020276 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P00568, Q9BVK9, Q9UQC7 | Gene names | AK1 | |||
|
Domain Architecture |
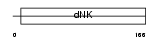
|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.029600 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
KAD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.016487 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6K8, Q96EC9 | Gene names | AK5 | |||
|
Domain Architecture |
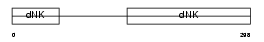
|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.031368 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
KAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.014488 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0Y5 | Gene names | Ak1 | |||
|
Domain Architecture |
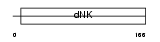
|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.001213 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
PEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.024970 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
SIA7B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.004552 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70277, Q3UWG0, Q9DC24 | Gene names | St6galnac2, Siat7, Siat7b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase II) (ST6GalNAc II) (Gal-beta-1,3-GalNAc alpha-2,6-sialyltransferase) (Sialyltransferase 7B). | |||||
|
SMBP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.039297 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.973099 (rank : 2) | θ value | 1.55827e-177 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.934407 (rank : 3) | θ value | 1.08726e-37 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.932859 (rank : 4) | θ value | 2.11701e-41 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.932689 (rank : 5) | θ value | 2.11701e-41 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.930482 (rank : 6) | θ value | 2.95404e-43 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.930417 (rank : 7) | θ value | 4.41421e-39 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.928370 (rank : 8) | θ value | 1.6774e-38 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.926465 (rank : 9) | θ value | 1.12253e-42 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.926410 (rank : 10) | θ value | 1.73182e-43 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.925799 (rank : 11) | θ value | 7.52953e-39 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.924862 (rank : 12) | θ value | 1.6774e-38 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.923518 (rank : 13) | θ value | 2.76489e-41 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.921950 (rank : 14) | θ value | 3.73686e-38 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.920831 (rank : 15) | θ value | 4.13158e-37 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.920345 (rank : 16) | θ value | 2.67802e-36 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.919950 (rank : 17) | θ value | 2.05049e-36 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.912214 (rank : 18) | θ value | 1.01765e-35 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.910141 (rank : 19) | θ value | 3.85808e-43 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.909873 (rank : 20) | θ value | 1.51363e-47 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.909736 (rank : 21) | θ value | 2.95404e-43 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.907324 (rank : 22) | θ value | 3.15612e-45 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.893274 (rank : 23) | θ value | 3.15612e-45 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.774610 (rank : 24) | θ value | 1.133e-10 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
PNKP_MOUSE
|
||||||
| NC score | 0.051368 (rank : 25) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLV6, Q6PFA3 | Gene names | Pnkp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
SMBP2_HUMAN
|
||||||
| NC score | 0.039297 (rank : 26) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38935, Q00443, Q14177 | Gene names | IGHMBP2, SMBP2, SMUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SMUBP-2 (EC 3.6.1.-) (ATP-dependent helicase IGHMBP2) (Immunoglobulin mu-binding protein 2) (SMUBP-2) (Glial factor 1) (GF-1). | |||||
|
PNKP_HUMAN
|
||||||
| NC score | 0.039242 (rank : 27) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96T60, Q9P1V2, Q9UKU8, Q9UNF8 | Gene names | PNKP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional polynucleotide phosphatase/kinase (Polynucleotide kinase- 3'-phosphatase) (DNA 5'-kinase/3'-phosphatase) [Includes: Polynucleotide 3'-phosphatase (EC 3.1.3.32) (2'(3')-polynucleotidase); Polynucleotide 5'-hydroxyl-kinase (EC 2.7.1.78)]. | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.031368 (rank : 28) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.029600 (rank : 29) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
PEO1_HUMAN
|
||||||
| NC score | 0.024970 (rank : 30) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.022697 (rank : 31) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
KAD1_HUMAN
|
||||||
| NC score | 0.020276 (rank : 32) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P00568, Q9BVK9, Q9UQC7 | Gene names | AK1 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
KAD5_HUMAN
|
||||||
| NC score | 0.016487 (rank : 33) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6K8, Q96EC9 | Gene names | AK5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 5 (EC 2.7.4.3) (ATP-AMP transphosphorylase). | |||||
|
HPS1_HUMAN
|
||||||
| NC score | 0.015623 (rank : 34) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92902, O15402, O15502 | Gene names | HPS1, HPS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 1 protein. | |||||
|
KAD1_MOUSE
|
||||||
| NC score | 0.014488 (rank : 35) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0Y5 | Gene names | Ak1 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.014475 (rank : 36) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.008050 (rank : 37) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SIA7B_MOUSE
|
||||||
| NC score | 0.004552 (rank : 38) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70277, Q3UWG0, Q9DC24 | Gene names | St6galnac2, Siat7, Siat7b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase II) (ST6GalNAc II) (Gal-beta-1,3-GalNAc alpha-2,6-sialyltransferase) (Sialyltransferase 7B). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.004317 (rank : 39) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
DSG4_HUMAN
|
||||||
| NC score | 0.003552 (rank : 40) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SJ6, Q6Y9L9, Q8IXV4 | Gene names | DSG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.001213 (rank : 41) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||