Please be patient as the page loads
|
PRP16_HUMAN
|
||||||
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHX16_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.943073 (rank : 10) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978906 (rank : 3) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX15_HUMAN
|
||||||
| θ value | 1.28067e-163 (rank : 4) | NC score | 0.978129 (rank : 4) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| θ value | 1.28067e-163 (rank : 5) | NC score | 0.978069 (rank : 5) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX35_HUMAN
|
||||||
| θ value | 4.71363e-158 (rank : 6) | NC score | 0.979024 (rank : 2) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX33_MOUSE
|
||||||
| θ value | 5.22361e-149 (rank : 7) | NC score | 0.976248 (rank : 6) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| θ value | 1.28662e-147 (rank : 8) | NC score | 0.975489 (rank : 7) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_MOUSE
|
||||||
| θ value | 2.35023e-141 (rank : 9) | NC score | 0.970966 (rank : 9) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX40_HUMAN
|
||||||
| θ value | 1.68428e-139 (rank : 10) | NC score | 0.971177 (rank : 8) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX34_MOUSE
|
||||||
| θ value | 3.32542e-95 (rank : 11) | NC score | 0.940768 (rank : 11) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX36_HUMAN
|
||||||
| θ value | 2.39424e-77 (rank : 12) | NC score | 0.909695 (rank : 15) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX29_HUMAN
|
||||||
| θ value | 2.92676e-75 (rank : 13) | NC score | 0.891835 (rank : 20) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX29_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 14) | NC score | 0.886012 (rank : 21) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX9_MOUSE
|
||||||
| θ value | 1.45253e-74 (rank : 15) | NC score | 0.904257 (rank : 17) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX30_HUMAN
|
||||||
| θ value | 3.24342e-66 (rank : 16) | NC score | 0.914005 (rank : 13) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| θ value | 5.53243e-66 (rank : 17) | NC score | 0.913456 (rank : 14) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX36_MOUSE
|
||||||
| θ value | 2.65945e-60 (rank : 18) | NC score | 0.903714 (rank : 18) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 5.7518e-47 (rank : 19) | NC score | 0.867369 (rank : 22) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 3.72821e-46 (rank : 20) | NC score | 0.907968 (rank : 16) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 2.41655e-45 (rank : 21) | NC score | 0.865491 (rank : 23) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX34_HUMAN
|
||||||
| θ value | 2.76489e-41 (rank : 22) | NC score | 0.923518 (rank : 12) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX9_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 23) | NC score | 0.899528 (rank : 19) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
YTDC2_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 24) | NC score | 0.736371 (rank : 24) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 25) | NC score | 0.064831 (rank : 28) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 26) | NC score | 0.071013 (rank : 26) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 27) | NC score | 0.047864 (rank : 37) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 28) | NC score | 0.057966 (rank : 31) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 29) | NC score | 0.098021 (rank : 25) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 30) | NC score | 0.051311 (rank : 35) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 31) | NC score | 0.051296 (rank : 36) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 32) | NC score | 0.038886 (rank : 48) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 33) | NC score | 0.027397 (rank : 62) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 34) | NC score | 0.046494 (rank : 40) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.030397 (rank : 56) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 36) | NC score | 0.064442 (rank : 29) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 37) | NC score | 0.066310 (rank : 27) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
DDX50_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.047259 (rank : 39) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.023920 (rank : 71) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DDX50_MOUSE
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.045582 (rank : 41) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.025352 (rank : 69) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.036588 (rank : 52) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.037901 (rank : 50) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.026042 (rank : 65) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.043485 (rank : 42) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
TTF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.038964 (rank : 47) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.016100 (rank : 89) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.006156 (rank : 114) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.034588 (rank : 53) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.027618 (rank : 61) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIL4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.022111 (rank : 72) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
RBY1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.013554 (rank : 91) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.038191 (rank : 49) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
COHA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.010000 (rank : 99) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
DDX46_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.032064 (rank : 55) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
DDX46_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.032737 (rank : 54) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.020186 (rank : 78) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.061783 (rank : 30) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.005550 (rank : 117) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.006793 (rank : 111) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.028134 (rank : 59) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.021478 (rank : 75) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.018310 (rank : 84) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.019641 (rank : 80) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SH24A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.018715 (rank : 83) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
TR150_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.047378 (rank : 38) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
FA50A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.009260 (rank : 102) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |
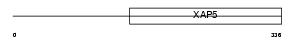
|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.041485 (rank : 44) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.041168 (rank : 45) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RAB7B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.001167 (rank : 130) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96AH8 | Gene names | RAB7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-7b. | |||||
|
RIPK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | -0.002501 (rank : 132) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
DDX28_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.018805 (rank : 82) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.004446 (rank : 121) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.005719 (rank : 115) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.019674 (rank : 79) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.025481 (rank : 68) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.028469 (rank : 58) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.020933 (rank : 77) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.021502 (rank : 74) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.021890 (rank : 73) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.025512 (rank : 67) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.007322 (rank : 110) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.039255 (rank : 46) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.026413 (rank : 64) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | -0.002536 (rank : 133) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | -0.003176 (rank : 134) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.037235 (rank : 51) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
ZFY2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | -0.003341 (rank : 135) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
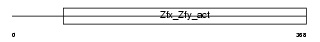
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
BAI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.001243 (rank : 129) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BOREA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.011559 (rank : 93) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHX3, Q6NVD3, Q8BHB1, Q8BHQ5, Q8BHQ9, Q9CRN4, Q9CS02, Q9CTF4 | Gene names | Cdca8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Borealin (Cell division cycle-associated protein 8) (MESrg). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.041503 (rank : 43) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
KAD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.011116 (rank : 96) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00568, Q9BVK9, Q9UQC7 | Gene names | AK1 | |||
|
Domain Architecture |
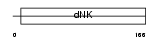
|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.004485 (rank : 120) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
PTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.006718 (rank : 113) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
S26A3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.004651 (rank : 119) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVC8 | Gene names | Slc26a3, Dra | |||
|
Domain Architecture |

|
|||||
| Description | Chloride anion exchanger (Protein DRA) (Down-regulated in adenoma) (Solute carrier family 26 member 3). | |||||
|
SEBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.012039 (rank : 92) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
SNPH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.007950 (rank : 105) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15079 | Gene names | SNPH, KIAA0374 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaphilin. | |||||
|
STK39_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | -0.003893 (rank : 137) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
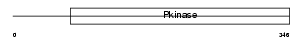
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.024695 (rank : 70) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.002733 (rank : 124) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
DDX47_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.021106 (rank : 76) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
DNJC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.017695 (rank : 87) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.015344 (rank : 90) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
FA38B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.006737 (rank : 112) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CD54, Q8BSM4 | Gene names | Fam38b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM38B. | |||||
|
GATB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.008217 (rank : 104) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75879, Q9P0S6, Q9Y2B8 | Gene names | PET112L, PET112 | |||
|
Domain Architecture |

|
|||||
| Description | Probable glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial precursor (EC 6.3.5.-) (Glu-ADT subunit B) (Cytochrome oxidase assembly factor PET112 homolog). | |||||
|
GIDRP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.007858 (rank : 106) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.010201 (rank : 98) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
LRRK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | -0.003674 (rank : 136) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.003022 (rank : 123) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NOP14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.028017 (rank : 60) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PP14C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.007689 (rank : 108) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4S0 | Gene names | Ppp1r14c, Kepi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14C (PKC-potentiated PP1 inhibitory protein) (Kinase-enhanced PP1 inhibitor). | |||||
|
RAB7B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.000172 (rank : 131) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEA8, Q8BJT0, Q8BZB1 | Gene names | Rab7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-7b. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.011125 (rank : 95) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.018035 (rank : 85) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.009278 (rank : 101) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.009293 (rank : 100) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SNUT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.011055 (rank : 97) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.027368 (rank : 63) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
B3GT4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.003991 (rank : 122) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96024 | Gene names | B3GALT4, GALT4 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1,3-galactosyltransferase 4 (EC 2.4.1.62) (Beta-1,3-GalTase 4) (Beta3Gal-T4) (b3Gal-T4) (Ganglioside galactosyltransferase) (UDP- galactose:beta-N-acetyl-galactosamine-beta-1,3-galactosyltransferase) (GAL-T2) (GalT4). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.028936 (rank : 57) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CAR10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.001821 (rank : 127) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.002703 (rank : 125) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.002197 (rank : 126) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.005572 (rank : 116) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.011371 (rank : 94) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
LSM11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.007735 (rank : 107) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P83369, Q7Z7P0, Q8N975 | Gene names | LSM11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U7 snRNA-associated Sm-like protein LSm11. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.019365 (rank : 81) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.005332 (rank : 118) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NIPA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.008273 (rank : 103) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.001605 (rank : 128) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.025598 (rank : 66) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.017392 (rank : 88) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.017772 (rank : 86) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TINF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.007410 (rank : 109) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSI4, Q9H904, Q9UHC2 | Gene names | TINF2, TIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TERF1-interacting nuclear factor 2 (TRF1-interacting nuclear protein 2). | |||||
|
ZBT24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | -0.004617 (rank : 138) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
NO40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.051950 (rank : 33) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NP64, Q6PKH4, Q9NYG4, Q9P0M8 | Gene names | ZCCHC17, PS1D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein) (Pnn-interacting nucleolar protein). | |||||
|
NO40_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051420 (rank : 34) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.055526 (rank : 32) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 135 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
DHX35_HUMAN
|
||||||
| NC score | 0.979024 (rank : 2) | θ value | 4.71363e-158 (rank : 6) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5Z1, Q5THR0, Q9H4H7, Q9H6T6 | Gene names | DHX35, C20orf15, DDX35 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX35 (EC 3.6.1.-) (DEAH box protein 35). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.978906 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
DHX15_HUMAN
|
||||||
| NC score | 0.978129 (rank : 4) | θ value | 1.28067e-163 (rank : 4) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43143, Q9NQT7 | Gene names | DHX15, DBP1, DDX15 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15) (ATP-dependent RNA helicase #46). | |||||
|
DHX15_MOUSE
|
||||||
| NC score | 0.978069 (rank : 5) | θ value | 1.28067e-163 (rank : 5) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35286, Q99L91 | Gene names | Dhx15, Ddx15, Deah9 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 (EC 3.6.1.-) (DEAH box protein 15). | |||||
|
DHX33_MOUSE
|
||||||
| NC score | 0.976248 (rank : 6) | θ value | 5.22361e-149 (rank : 7) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q80VY9, Q8BS50 | Gene names | Dhx33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX33_HUMAN
|
||||||
| NC score | 0.975489 (rank : 7) | θ value | 1.28662e-147 (rank : 8) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H6R0, Q4G149, Q5CZ73, Q9H5M9 | Gene names | DHX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX33 (EC 3.6.1.-) (DEAH box protein 33). | |||||
|
DHX40_HUMAN
|
||||||
| NC score | 0.971177 (rank : 8) | θ value | 1.68428e-139 (rank : 10) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IX18, Q5JPH4, Q8TC86, Q8WY53, Q9BXM1, Q9H6M9 | Gene names | DHX40, DDX40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40) (ARG147) (Protein PAD). | |||||
|
DHX40_MOUSE
|
||||||
| NC score | 0.970966 (rank : 9) | θ value | 2.35023e-141 (rank : 9) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PE54, Q8BPH2, Q8CD88, Q9CWN3 | Gene names | Dhx40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX40 (EC 3.6.1.-) (DEAH box protein 40). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.943073 (rank : 10) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
DHX34_MOUSE
|
||||||
| NC score | 0.940768 (rank : 11) | θ value | 3.32542e-95 (rank : 11) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBV3 | Gene names | Dhx34, Ddx34, Kiaa0134 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX34_HUMAN
|
||||||
| NC score | 0.923518 (rank : 12) | θ value | 2.76489e-41 (rank : 22) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14147 | Gene names | DHX34, DDX34, KIAA0134 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX34 (EC 3.6.1.-) (DEAH box protein 34). | |||||
|
DHX30_HUMAN
|
||||||
| NC score | 0.914005 (rank : 13) | θ value | 3.24342e-66 (rank : 16) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7L2E3, O94965, Q7Z753, Q96CH4 | Gene names | DHX30, KIAA0890 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX30_MOUSE
|
||||||
| NC score | 0.913456 (rank : 14) | θ value | 5.53243e-66 (rank : 17) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99PU8, Q3U4Z4, Q3UFA0, Q3UJS4, Q91WA7, Q99KN7 | Gene names | Dhx30, Helg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX30 (EC 3.6.1.-) (DEAH box protein 30). | |||||
|
DHX36_HUMAN
|
||||||
| NC score | 0.909695 (rank : 15) | θ value | 2.39424e-77 (rank : 12) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H2U1, Q70JU3, Q8IYE5, Q9P240 | Gene names | DHX36, DDX36, KIAA1488, MLEL1, RHAU | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.907968 (rank : 16) | θ value | 3.72821e-46 (rank : 20) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
DHX9_MOUSE
|
||||||
| NC score | 0.904257 (rank : 17) | θ value | 1.45253e-74 (rank : 15) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70133, O35931, O54703, Q5FWY1, Q6R5F7, Q9CSA2 | Gene names | Dhx9, Ddx9 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9) (mHEL-5). | |||||
|
DHX36_MOUSE
|
||||||
| NC score | 0.903714 (rank : 18) | θ value | 2.65945e-60 (rank : 18) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VHK9, Q6ZPP7, Q9CSE8 | Gene names | Dhx36, Ddx36, Kiaa1488, Mlel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DHX36 (EC 3.6.1.-) (DEAH box protein 36) (MLE-like protein 1) (RNA helicase associated with AU-rich element ARE). | |||||
|
DHX9_HUMAN
|
||||||
| NC score | 0.899528 (rank : 19) | θ value | 2.86122e-38 (rank : 23) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q08211, Q5VY62, Q6PD69, Q99556 | Gene names | DHX9, DDX9, NDH2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase A (EC 3.6.1.-) (Nuclear DNA helicase II) (NDH II) (DEAH box protein 9). | |||||
|
DHX29_HUMAN
|
||||||
| NC score | 0.891835 (rank : 20) | θ value | 2.92676e-75 (rank : 13) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z478, O75549, Q63HN0, Q63HN3, Q8IWW2, Q8N3A1, Q9UMH2 | Gene names | DHX29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29) (Nucleic acid helicase DDXx). | |||||
|
DHX29_MOUSE
|
||||||
| NC score | 0.886012 (rank : 21) | θ value | 4.99229e-75 (rank : 14) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PGC1, Q8BQJ4, Q8BT01, Q8C9B7, Q8C9H9 | Gene names | Dhx29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX29 (EC 3.6.1.-) (DEAH box protein 29). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.867369 (rank : 22) | θ value | 5.7518e-47 (rank : 19) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.865491 (rank : 23) | θ value | 2.41655e-45 (rank : 21) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
YTDC2_HUMAN
|
||||||
| NC score | 0.736371 (rank : 24) | θ value | 1.17247e-07 (rank : 24) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H6S0 | Gene names | YTHDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain-containing protein 2. | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.098021 (rank : 25) | θ value | 0.00390308 (rank : 29) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.071013 (rank : 26) | θ value | 0.000270298 (rank : 26) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.066310 (rank : 27) | θ value | 0.0736092 (rank : 37) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.064831 (rank : 28) | θ value | 0.000121331 (rank : 25) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.064442 (rank : 29) | θ value | 0.0736092 (rank : 36) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.061783 (rank : 30) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.057966 (rank : 31) | θ value | 0.000786445 (rank : 28) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.055526 (rank : 32) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
NO40_HUMAN
|
||||||
| NC score | 0.051950 (rank : 33) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NP64, Q6PKH4, Q9NYG4, Q9P0M8 | Gene names | ZCCHC17, PS1D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein) (Pnn-interacting nucleolar protein). | |||||
|
NO40_MOUSE
|
||||||
| NC score | 0.051420 (rank : 34) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.051311 (rank : 35) | θ value | 0.00665767 (rank : 30) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.051296 (rank : 36) | θ value | 0.00665767 (rank : 31) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.047864 (rank : 37) | θ value | 0.000461057 (rank : 27) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.047378 (rank : 38) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
DDX50_HUMAN
|
||||||
| NC score | 0.047259 (rank : 39) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQ39, Q8WV76, Q9BWI8 | Gene names | DDX50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.046494 (rank : 40) | θ value | 0.0148317 (rank : 34) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
DDX50_MOUSE
|
||||||
| NC score | 0.045582 (rank : 41) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99MJ9 | Gene names | Ddx50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX50 (EC 3.6.1.-) (DEAD box protein 50) (Nucleolar protein Gu2) (Gu-beta). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.043485 (rank : 42) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.041503 (rank : 43) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.041485 (rank : 44) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.041168 (rank : 45) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.039255 (rank : 46) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
TTF1_MOUSE
|
||||||
| NC score | 0.038964 (rank : 47) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62187, Q9JKK5 | Gene names | Ttf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 1 (TTF-1) (TTF-I) (mTFF-I) (RNA polymerase I termination factor). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.038886 (rank : 48) | θ value | 0.0113563 (rank : 32) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.038191 (rank : 49) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.037901 (rank : 50) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.037235 (rank : 51) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.036588 (rank : 52) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.034588 (rank : 53) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
DDX46_MOUSE
|
||||||
| NC score | 0.032737 (rank : 54) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q569Z5, Q6ZQ42, Q8R0R6 | Gene names | Ddx46, Kiaa0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46). | |||||
|
DDX46_HUMAN
|
||||||
| NC score | 0.032064 (rank : 55) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7L014, O94894, Q96EI0, Q9Y658 | Gene names | DDX46, KIAA0801 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX46 (EC 3.6.1.-) (DEAD box protein 46) (PRP5 homolog). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.030397 (rank : 56) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.028936 (rank : 57) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.028469 (rank : 58) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.028134 (rank : 59) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
NOP14_HUMAN
|
||||||
| NC score | 0.028017 (rank : 60) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P78316, Q7LGI5, Q7Z6K0, Q8TBR6 | Gene names | NOP14, C4orf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.027618 (rank : 61) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.027397 (rank : 62) | θ value | 0.0148317 (rank : 33) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.027368 (rank : 63) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
DDX28_HUMAN
|
||||||
| NC score | 0.026413 (rank : 64) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NUL7 | Gene names | DDX28, MDDX28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.026042 (rank : 65) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.025598 (rank : 66) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.025512 (rank : 67) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.025481 (rank : 68) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.025352 (rank : 69) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.024695 (rank : 70) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.023920 (rank : 71) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PPIL4_HUMAN
|
||||||
| NC score | 0.022111 (rank : 72) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WUA2, Q7Z3Q5 | Gene names | PPIL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peptidyl-prolyl cis-trans isomerase-like 4 (EC 5.2.1.8) (PPIase) (Rotamase) (Cyclophilin-like protein PPIL4). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.021890 (rank : 73) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.021502 (rank : 74) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.021478 (rank : 75) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
DDX47_MOUSE
|
||||||
| NC score | 0.021106 (rank : 76) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CWX9 | Gene names | Ddx47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX47 (EC 3.6.1.-) (DEAD box protein 47). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.020933 (rank : 77) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.020186 (rank : 78) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.019674 (rank : 79) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.019641 (rank : 80) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.019365 (rank : 81) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
DDX28_MOUSE
|
||||||
| NC score | 0.018805 (rank : 82) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CWT6, Q3TQM0 | Gene names | Ddx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX28 (EC 3.6.1.-) (Mitochondrial DEAD box protein 28). | |||||
|
SH24A_MOUSE
|
||||||
| NC score | 0.018715 (rank : 83) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.018310 (rank : 84) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.018035 (rank : 85) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.017772 (rank : 86) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
DNJC8_HUMAN
|
||||||
| NC score | 0.017695 (rank : 87) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75937, Q6IBA4, Q8N4Z5, Q9P051, Q9P067 | Gene names | DNAJC8, SPF31 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 8 (Splicing protein spf31). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.017392 (rank : 88) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.016100 (rank : 89) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.015344 (rank : 90) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
RBY1A_HUMAN
|
||||||
| NC score | 0.013554 (rank : 91) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15414, Q15377, Q8NHR0 | Gene names | RBMY1A1, RBM1, YRRM1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, Y chromosome, family 1 member A1 (RNA- binding motif protein 1). | |||||
|
SEBP2_HUMAN
|
||||||
| NC score | 0.012039 (rank : 92) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
BOREA_MOUSE
|
||||||
| NC score | 0.011559 (rank : 93) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHX3, Q6NVD3, Q8BHB1, Q8BHQ5, Q8BHQ9, Q9CRN4, Q9CS02, Q9CTF4 | Gene names | Cdca8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Borealin (Cell division cycle-associated protein 8) (MESrg). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.011371 (rank : 94) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.011125 (rank : 95) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
KAD1_HUMAN
|
||||||
| NC score | 0.011116 (rank : 96) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00568, Q9BVK9, Q9UQC7 | Gene names | AK1 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate kinase isoenzyme 1 (EC 2.7.4.3) (ATP-AMP transphosphorylase) (AK1) (Myokinase). | |||||
|
SNUT1_HUMAN
|
||||||
| NC score | 0.011055 (rank : 97) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.010201 (rank : 98) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
COHA1_HUMAN
|
||||||
| NC score | 0.010000 (rank : 99) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UMD9, Q02802, Q99018, Q9NQK9, Q9UC14 | Gene names | COL17A1, BP180, BPAG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.009293 (rank : 100) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.009278 (rank : 101) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
FA50A_MOUSE
|
||||||
| NC score | 0.009260 (rank : 102) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV03, Q9D866 | Gene names | Fam50a, D0HXS9928E, Xap5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAM50A (XAP-5 protein). | |||||
|
NIPA_MOUSE
|
||||||
| NC score | 0.008273 (rank : 103) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
GATB_HUMAN
|
||||||
| NC score | 0.008217 (rank : 104) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75879, Q9P0S6, Q9Y2B8 | Gene names | PET112L, PET112 | |||
|
Domain Architecture |

|
|||||
| Description | Probable glutamyl-tRNA(Gln) amidotransferase subunit B, mitochondrial precursor (EC 6.3.5.-) (Glu-ADT subunit B) (Cytochrome oxidase assembly factor PET112 homolog). | |||||
|
SNPH_HUMAN
|
||||||
| NC score | 0.007950 (rank : 105) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15079 | Gene names | SNPH, KIAA0374 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaphilin. | |||||
|
GIDRP_MOUSE
|
||||||
| NC score | 0.007858 (rank : 106) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
LSM11_HUMAN
|
||||||
| NC score | 0.007735 (rank : 107) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P83369, Q7Z7P0, Q8N975 | Gene names | LSM11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U7 snRNA-associated Sm-like protein LSm11. | |||||
|
PP14C_MOUSE
|
||||||
| NC score | 0.007689 (rank : 108) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4S0 | Gene names | Ppp1r14c, Kepi | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase 1 regulatory subunit 14C (PKC-potentiated PP1 inhibitory protein) (Kinase-enhanced PP1 inhibitor). | |||||
|
TINF2_HUMAN
|
||||||
| NC score | 0.007410 (rank : 109) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BSI4, Q9H904, Q9UHC2 | Gene names | TINF2, TIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TERF1-interacting nuclear factor 2 (TRF1-interacting nuclear protein 2). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.007322 (rank : 110) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.006793 (rank : 111) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
FA38B_MOUSE
|
||||||
| NC score | 0.006737 (rank : 112) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CD54, Q8BSM4 | Gene names | Fam38b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM38B. | |||||
|
PTC1_MOUSE
|
||||||
| NC score | 0.006718 (rank : 113) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.006156 (rank : 114) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.005719 (rank : 115) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.005572 (rank : 116) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.005550 (rank : 117) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.005332 (rank : 118) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
S26A3_MOUSE
|
||||||
| NC score | 0.004651 (rank : 119) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVC8 | Gene names | Slc26a3, Dra | |||
|
Domain Architecture |

|
|||||
| Description | Chloride anion exchanger (Protein DRA) (Down-regulated in adenoma) (Solute carrier family 26 member 3). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.004485 (rank : 120) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.004446 (rank : 121) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
B3GT4_HUMAN
|
||||||
| NC score | 0.003991 (rank : 122) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96024 | Gene names | B3GALT4, GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-galactosyltransferase 4 (EC 2.4.1.62) (Beta-1,3-GalTase 4) (Beta3Gal-T4) (b3Gal-T4) (Ganglioside galactosyltransferase) (UDP- galactose:beta-N-acetyl-galactosamine-beta-1,3-galactosyltransferase) (GAL-T2) (GalT4). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.003022 (rank : 123) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.002733 (rank : 124) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.002703 (rank : 125) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.002197 (rank : 126) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
CAR10_HUMAN
|
||||||
| NC score | 0.001821 (rank : 127) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.001605 (rank : 128) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BAI2_HUMAN
|
||||||
| NC score | 0.001243 (rank : 129) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
RAB7B_HUMAN
|
||||||
| NC score | 0.001167 (rank : 130) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96AH8 | Gene names | RAB7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-7b. | |||||
|
RAB7B_MOUSE
|
||||||
| NC score | 0.000172 (rank : 131) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEA8, Q8BJT0, Q8BZB1 | Gene names | Rab7b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-7b. | |||||
|
RIPK1_MOUSE
|
||||||
| NC score | -0.002501 (rank : 132) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
GLI3_MOUSE
|
||||||
| NC score | -0.002536 (rank : 133) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | -0.003176 (rank : 134) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZFY2_MOUSE
|
||||||
| NC score | -0.003341 (rank : 135) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
LRRK2_HUMAN
|
||||||
| NC score | -0.003674 (rank : 136) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||
|
STK39_MOUSE
|
||||||
| NC score | -0.003893 (rank : 137) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
ZBT24_HUMAN
|
||||||
| NC score | -0.004617 (rank : 138) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 135 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||