Please be patient as the page loads
|
GIDRP_MOUSE
|
||||||
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GIDRP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.960006 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z5L2, O60598, Q5W0B4, Q5W0B5, Q86VT9, Q8N9H0 | Gene names | GIDRP88, C10orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 (Putative mitochondrial space protein 32.1). | |||||
|
GIDRP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 3) | NC score | 0.218721 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 4) | NC score | 0.179038 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PARN_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.161941 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95453 | Gene names | PARN, DAN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A)-specific ribonuclease PARN (EC 3.1.13.4) (Polyadenylate- specific ribonuclease) (Deadenylating nuclease) (Deadenylation nuclease). | |||||
|
PARN_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.156626 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDG3, Q8C7N6, Q9DC46 | Gene names | Parn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A)-specific ribonuclease PARN (EC 3.1.13.4) (Polyadenylate- specific ribonuclease). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.120058 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.027162 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.087816 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.060985 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TM16C_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.042721 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYT9 | Gene names | TMEM16C, C11orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 16C. | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.034275 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
ANLN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.055720 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.018980 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
NF2L3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.043965 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
RB15B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.054550 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.041880 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.039809 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
K0146_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.065264 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.030352 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.024798 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.026338 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.030871 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.050247 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.050509 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.058215 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.034384 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.098598 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.048716 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
SMBT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.025459 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.032389 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.002204 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.069754 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.024715 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
NO40_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.041746 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.029365 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.036729 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
DCAK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.005629 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLM8, Q6P207 | Gene names | Dcamkl1, Dclk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
HMGA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.064323 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52927, Q3UQW0 | Gene names | Hmga2, Hmgic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.044727 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.032049 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.007476 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CO5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.015533 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
DCAK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.005310 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15075 | Gene names | DCAMKL1, KIAA0369 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.022331 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
NEDD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.018807 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P33215, Q6NXV3, Q8BN12, Q8BN86, Q8BQL9, Q9CWK2 | Gene names | Nedd1, Nedd-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NEDD1 (Neural precursor cell expressed developmentally down- regulated protein 1). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.008238 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
OLIG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.030401 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |
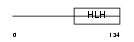
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
RB15B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.041812 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
ZN439_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.000953 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NDP4, Q8IYZ7, Q96SU1 | Gene names | ZNF439 | |||
|
Domain Architecture |
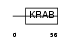
|
|||||
| Description | Zinc finger protein 439. | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.025567 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.026852 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
EST1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.020992 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.008056 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.008071 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
RT05_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.041498 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N87 | Gene names | Mrps5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 28S ribosomal protein S5 (S5mt) (MRP-S5). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.036878 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.025236 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.058426 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AF10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.012940 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.015462 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CD008_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.039259 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
CROP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.036272 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
CROP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.036668 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
GLCM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.022249 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17439 | Gene names | Gba | |||
|
Domain Architecture |

|
|||||
| Description | Glucosylceramidase precursor (EC 3.2.1.45) (Beta-glucocerebrosidase) (Acid beta-glucosidase) (D-glucosyl-N-acylsphingosine glucohydrolase). | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.038956 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.018268 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.035199 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.058491 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.023666 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.023668 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.031097 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
XE7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.014364 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.025335 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.017169 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.006694 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
HMGA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.050108 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52926 | Gene names | HMGA2, HMGIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
MDM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.024226 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00987, Q13226, Q13297, Q13298, Q13299, Q13300, Q13301, Q53XW0, Q71TW9, Q9UGI3, Q9UMT8 | Gene names | MDM2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein) (Hdm2). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.004247 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.029975 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.010968 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
PRP16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.007858 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
RUSC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.013737 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
SPESP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.023461 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UW49, Q8NG22, Q8WVH8 | Gene names | SPESP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm equatorial segment protein 1 precursor (SP-ESP) (Equatorial segment protein) (ESP) (Glycosylated 38 kDa sperm protein C-7/8). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.025422 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.026540 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CB027_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.025113 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q580R0, O43575, Q52M10, Q86XG2 | Gene names | C2orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf27. | |||||
|
CUBN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.005641 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DMXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.008909 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
FBX30_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.014105 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TB52, Q9BXZ7 | Gene names | FBXO30, FBX30 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 30. | |||||
|
MUC13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.017292 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.007915 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.010526 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RAD52_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.019001 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43351, Q13205 | Gene names | RAD52 | |||
|
Domain Architecture |
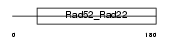
|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.022208 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
ZN692_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.001081 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053920 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.061065 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.055375 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.054836 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SG16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050306 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
GIDRP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
GIDRP_HUMAN
|
||||||
| NC score | 0.960006 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7Z5L2, O60598, Q5W0B4, Q5W0B5, Q86VT9, Q8N9H0 | Gene names | GIDRP88, C10orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 (Putative mitochondrial space protein 32.1). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.218721 (rank : 3) | θ value | 1.38499e-08 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.179038 (rank : 4) | θ value | 1.29631e-06 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PARN_HUMAN
|
||||||
| NC score | 0.161941 (rank : 5) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95453 | Gene names | PARN, DAN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A)-specific ribonuclease PARN (EC 3.1.13.4) (Polyadenylate- specific ribonuclease) (Deadenylating nuclease) (Deadenylation nuclease). | |||||
|
PARN_MOUSE
|
||||||
| NC score | 0.156626 (rank : 6) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDG3, Q8C7N6, Q9DC46 | Gene names | Parn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A)-specific ribonuclease PARN (EC 3.1.13.4) (Polyadenylate- specific ribonuclease). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.120058 (rank : 7) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.098598 (rank : 8) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.087816 (rank : 9) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.069754 (rank : 10) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
K0146_MOUSE
|
||||||
| NC score | 0.065264 (rank : 11) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGX7, Q3TCD0, Q5U457, Q6A0B6, Q6IS60, Q811E1, Q8BWD6, Q8R305 | Gene names | Kiaa0146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0146. | |||||
|
HMGA2_MOUSE
|
||||||
| NC score | 0.064323 (rank : 12) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52927, Q3UQW0 | Gene names | Hmga2, Hmgic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.061065 (rank : 13) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.060985 (rank : 14) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.058491 (rank : 15) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.058426 (rank : 16) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.058215 (rank : 17) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.055720 (rank : 18) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.055375 (rank : 19) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.054836 (rank : 20) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RB15B_MOUSE
|
||||||
| NC score | 0.054550 (rank : 21) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PHZ5, Q8C6G2 | Gene names | Rbm15b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.053920 (rank : 22) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.050509 (rank : 23) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
SG16_MOUSE
|
||||||
| NC score | 0.050306 (rank : 24) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02815, Q62263 | Gene names | Spt1, Spt-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 16.5 kDa submandibular gland glycoprotein precursor (Salivary protein 1). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.050247 (rank : 25) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
HMGA2_HUMAN
|
||||||
| NC score | 0.050108 (rank : 26) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P52926 | Gene names | HMGA2, HMGIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMGI-C (High mobility group AT-hook protein 2). | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.048716 (rank : 27) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.044727 (rank : 28) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
NF2L3_MOUSE
|
||||||
| NC score | 0.043965 (rank : 29) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WTM4 | Gene names | Nfe2l3, Nrf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
TM16C_HUMAN
|
||||||
| NC score | 0.042721 (rank : 30) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYT9 | Gene names | TMEM16C, C11orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protein 16C. | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.041880 (rank : 31) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
RB15B_HUMAN
|
||||||
| NC score | 0.041812 (rank : 32) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDT2, Q6QE19, Q9BV96 | Gene names | RBM15B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative RNA-binding protein 15B (RNA-binding motif protein 15B). | |||||
|
NO40_MOUSE
|
||||||
| NC score | 0.041746 (rank : 33) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
RT05_MOUSE
|
||||||
| NC score | 0.041498 (rank : 34) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N87 | Gene names | Mrps5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 28S ribosomal protein S5 (S5mt) (MRP-S5). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.039809 (rank : 35) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.039259 (rank : 36) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.038956 (rank : 37) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.036878 (rank : 38) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.036729 (rank : 39) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.036668 (rank : 40) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
CROP_HUMAN
|
||||||
| NC score | 0.036272 (rank : 41) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95232, Q6PHR9, Q9NUY0, Q9P2S7 | Gene names | CROP, CREAP1, O48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein (cAMP regulatory element-associated protein 1) (CRE-associated protein 1) (CREAP-1) (Luc7A) (Okadaic acid-inducible phosphoprotein OA48-18). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.035199 (rank : 42) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.034384 (rank : 43) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.034275 (rank : 44) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.032389 (rank : 45) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.032049 (rank : 46) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.031097 (rank : 47) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.030871 (rank : 48) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
OLIG1_MOUSE
|
||||||
| NC score | 0.030401 (rank : 49) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.030352 (rank : 50) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.029975 (rank : 51) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.029365 (rank : 52) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.027162 (rank : 53) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.026852 (rank : 54) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.026540 (rank : 55) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.026338 (rank : 56) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.025567 (rank : 57) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
SMBT2_MOUSE
|
||||||
| NC score | 0.025459 (rank : 58) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5DTW2, Q80VG7 | Gene names | Sfmbt2, Kiaa1617 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scm-like with four MBT domains protein 2. | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.025422 (rank : 59) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.025335 (rank : 60) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.025236 (rank : 61) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
CB027_HUMAN
|
||||||
| NC score | 0.025113 (rank : 62) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q580R0, O43575, Q52M10, Q86XG2 | Gene names | C2orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf27. | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.024798 (rank : 63) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.024715 (rank : 64) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
MDM2_HUMAN
|
||||||
| NC score | 0.024226 (rank : 65) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00987, Q13226, Q13297, Q13298, Q13299, Q13300, Q13301, Q53XW0, Q71TW9, Q9UGI3, Q9UMT8 | Gene names | MDM2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3 Mdm2 (EC 6.3.2.-) (p53-binding protein Mdm2) (Oncoprotein Mdm2) (Double minute 2 protein) (Hdm2). | |||||
|
TCAL5_MOUSE
|
||||||
| NC score | 0.023668 (rank : 66) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.023666 (rank : 67) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
SPESP_HUMAN
|
||||||
| NC score | 0.023461 (rank : 68) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UW49, Q8NG22, Q8WVH8 | Gene names | SPESP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm equatorial segment protein 1 precursor (SP-ESP) (Equatorial segment protein) (ESP) (Glycosylated 38 kDa sperm protein C-7/8). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.022331 (rank : 69) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
GLCM_MOUSE
|
||||||
| NC score | 0.022249 (rank : 70) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17439 | Gene names | Gba | |||
|
Domain Architecture |

|
|||||
| Description | Glucosylceramidase precursor (EC 3.2.1.45) (Beta-glucocerebrosidase) (Acid beta-glucosidase) (D-glucosyl-N-acylsphingosine glucohydrolase). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.022208 (rank : 71) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
EST1A_HUMAN
|
||||||
| NC score | 0.020992 (rank : 72) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86US8, O94837, Q86VH6, Q9UF60 | Gene names | EST1A, C17orf31, KIAA0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A) (hSmg5/7a). | |||||
|
RAD52_HUMAN
|
||||||
| NC score | 0.019001 (rank : 73) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43351, Q13205 | Gene names | RAD52 | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair protein RAD52 homolog. | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.018980 (rank : 74) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
NEDD1_MOUSE
|
||||||
| NC score | 0.018807 (rank : 75) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P33215, Q6NXV3, Q8BN12, Q8BN86, Q8BQL9, Q9CWK2 | Gene names | Nedd1, Nedd-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NEDD1 (Neural precursor cell expressed developmentally down- regulated protein 1). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.018268 (rank : 76) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MUC13_HUMAN
|
||||||
| NC score | 0.017292 (rank : 77) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H3R2, Q6UWD9, Q9NXT5 | Gene names | MUC13, DRCC1, RECC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-13 precursor (Down-regulated in colon cancer 1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017169 (rank : 78) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CO5_MOUSE
|
||||||
| NC score | 0.015533 (rank : 79) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06684 | Gene names | C5, Hc | |||
|
Domain Architecture |

|
|||||
| Description | Complement C5 precursor (Hemolytic complement) [Contains: Complement C5 beta chain; Complement C5 alpha chain; C5a anaphylatoxin; Complement C5 alpha' chain]. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.015462 (rank : 80) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.014364 (rank : 81) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
FBX30_HUMAN
|
||||||
| NC score | 0.014105 (rank : 82) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TB52, Q9BXZ7 | Gene names | FBXO30, FBX30 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 30. | |||||
|
RUSC1_MOUSE
|
||||||
| NC score | 0.013737 (rank : 83) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.012940 (rank : 84) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.010968 (rank : 85) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.010526 (rank : 86) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DMXL1_MOUSE
|
||||||
| NC score | 0.008909 (rank : 87) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.008238 (rank : 88) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.008071 (rank : 89) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.008056 (rank : 90) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.007915 (rank : 91) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PRP16_HUMAN
|
||||||
| NC score | 0.007858 (rank : 92) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92620, O75212, Q96HN7 | Gene names | DHX38, DDX38, KIAA0224, PRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase DHX38) (DEAH box protein 38). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.007476 (rank : 93) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.006694 (rank : 94) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.005641 (rank : 95) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DCAK1_MOUSE
|
||||||
| NC score | 0.005629 (rank : 96) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JLM8, Q6P207 | Gene names | Dcamkl1, Dclk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
DCAK1_HUMAN
|
||||||
| NC score | 0.005310 (rank : 97) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15075 | Gene names | DCAMKL1, KIAA0369 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase DCAMKL1 (EC 2.7.11.1) (Doublecortin- like and CAM kinase-like 1). | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.004247 (rank : 98) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.002204 (rank : 99) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
ZN692_MOUSE
|
||||||
| NC score | 0.001081 (rank : 100) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
ZN439_HUMAN
|
||||||
| NC score | 0.000953 (rank : 101) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NDP4, Q8IYZ7, Q96SU1 | Gene names | ZNF439 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 439. | |||||