Please be patient as the page loads
|
TDRD6_MOUSE
|
||||||
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TDRD6_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
TDRD1_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 2) | NC score | 0.673811 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD1_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 3) | NC score | 0.663018 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD7_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 4) | NC score | 0.455819 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD5_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.503809 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRKH_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 6) | NC score | 0.413135 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRD7_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 7) | NC score | 0.440349 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 8) | NC score | 0.407728 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
SND1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 9) | NC score | 0.394993 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7KZF4, Q13122 | Gene names | SND1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator) (EBNA2 coactivator p100). | |||||
|
SND1_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 10) | NC score | 0.391368 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q78PY7, Q3TT46, Q922L5, Q9R0S1 | Gene names | Snd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator). | |||||
|
STK31_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 11) | NC score | 0.215161 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MW1 | Gene names | Stk31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1). | |||||
|
STK31_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 12) | NC score | 0.210708 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXU1, Q9BXH8 | Gene names | STK31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1) (Serine/threonine- protein kinase NYD-SPK). | |||||
|
TDRD4_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 13) | NC score | 0.389504 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
AKAP1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.274732 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.265599 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
TDRD3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.151390 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
RNF17_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.123552 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXT8, Q9BXT7 | Gene names | RNF17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
RNF17_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.112474 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MV7 | Gene names | Rnf17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
TDRD3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.141442 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91W18, Q8BZW6 | Gene names | Tdrd3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
BRD3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.032955 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.030133 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
LYPA1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.057249 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97823, Q3TJZ0, Q7TPX1, Q8BWM6 | Gene names | Lypla1, Apt1, Pla1a | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-protein thioesterase 1 (EC 3.1.2.-) (Lysophospholipase 1) (Lysophospholipase I) (LysoPLA I). | |||||
|
CJ006_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.047493 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX21, Q5W0L8, Q9NPE8 | Gene names | C10orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6. | |||||
|
SPF30_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.122032 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
FAS_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.043746 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
LYPA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.051638 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75608, O43202, Q9UQF9 | Gene names | LYPLA1, APT1, LPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-protein thioesterase 1 (EC 3.1.2.-) (Lysophospholipase 1) (Lysophospholipase I). | |||||
|
SPA12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.011368 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMF5, Q9CQ32 | Gene names | Serpina12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin). | |||||
|
SPF30_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.115763 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.029431 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.013188 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.030632 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SCTM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.059501 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVN6, O00466 | Gene names | SECTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted and transmembrane protein 1 precursor (Protein K12). | |||||
|
FA10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.007528 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
GDPD2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.025824 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESM6 | Gene names | Gdpd2, Gde3, Obdpf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 2 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 3) (Osteoblast differentiation promoting factor). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.019499 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
WNK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.004504 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
CSKI2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.009401 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.027135 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
FA8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.016118 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
FAS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.039594 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
PTPRD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.008646 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
SNTB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.020356 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13884, O14912, Q4KMG8 | Gene names | SNTB1, SNT2B1 | |||
|
Domain Architecture |
|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Tax interaction protein 43) (TIP-43) (Syntrophin 2) (BSYN2). | |||||
|
APLP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.022894 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.008298 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
GIDRP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.025236 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
PB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.022241 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
TTC16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.024309 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
U384_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.038210 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3C1, Q6IAI6, Q8IXL5 | Gene names | ames=CGI-117, HSPC111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0384 protein CGI-117 (HBV pre-S2 trans-regulated protein 3). | |||||
|
ACET_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.019938 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22966 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, testis-specific isoform precursor (EC 3.4.15.1) (EC 3.2.1.-) (ACE-T) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin-converting enzyme, testis- specific isoform, soluble form]. | |||||
|
ACE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.019521 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12821, Q53YX9, Q59GY8 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) (CD143 antigen) [Contains: Angiotensin-converting enzyme, somatic isoform, soluble form]. | |||||
|
F13A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.010624 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH61, Q3TNF9, Q8BIP2 | Gene names | F13a1, F13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coagulation factor XIII A chain precursor (EC 2.3.2.13) (Coagulation factor XIIIa) (Protein-glutamine gamma-glutamyltransferase A chain) (Transglutaminase A chain). | |||||
|
FBLN4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.007656 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVJ9 | Gene names | Efemp2, Fbln4, Mbp1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (Mutant p53-binding protein 1). | |||||
|
FBX38_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.022402 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.020868 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.007763 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.018451 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.013718 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.007636 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CP1A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.004441 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04798 | Gene names | CYP1A1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1) (P450 form 6) (P450-C). | |||||
|
F10A5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.012678 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
HMGN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.026444 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCB1, Q7M737, Q99JU1 | Gene names | Hmgn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group nucleosome-binding domain-containing protein 3. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.004281 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NUDT4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.017278 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZJ9, Q53EZ2, Q68DD7, Q9NPC5, Q9NS30, Q9NZK0, Q9NZK1 | Gene names | NUDT4, DIPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diphosphoinositol polyphosphate phosphohydrolase 2 (EC 3.6.1.52) (DIPP-2) (Diadenosine 5',5'''-P1,P6-hexaphosphate hydrolase 2) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 4) (Nudix motif 4). | |||||
|
NUDT4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.017254 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2U6, Q8BXB9, Q9D3T6 | Gene names | Nudt4, Dipp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diphosphoinositol polyphosphate phosphohydrolase 2 (EC 3.6.1.52) (DIPP-2) (Diadenosine 5',5'''-P1,P6-hexaphosphate hydrolase 2) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 4) (Nudix motif 4). | |||||
|
PASK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.000515 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||
|
AQR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.011934 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.011606 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-G2. | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.016255 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
EPMIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.015095 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7L775, O94866, Q9H3L3 | Gene names | EPM2AIP1, KIAA0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
FBLN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.006272 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
GP124_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.004571 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
IRX5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.007825 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.004490 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
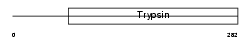
KLK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.003437 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y337, Q9HBG8 | Gene names | KLK5, SCTE | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-5 precursor (EC 3.4.21.-) (Stratum corneum tryptic enzyme) (Kallikrein-like protein 2) (KLK-L2). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.011912 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.011730 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PAX5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.010111 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02548, O75933 | Gene names | PAX5 | |||
|
Domain Architecture |
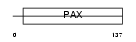
|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PAX5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.010103 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02650 | Gene names | Pax5, Pax-5 | |||
|
Domain Architecture |
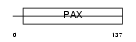
|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.010568 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.016175 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.016461 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SNTB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.015391 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99L88, O35925 | Gene names | Sntb1, Snt2b1 | |||
|
Domain Architecture |
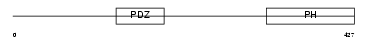
|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Syntrophin 2). | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
TDRD1_MOUSE
|
||||||
| NC score | 0.673811 (rank : 2) | θ value | 3.74554e-30 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD1_HUMAN
|
||||||
| NC score | 0.663018 (rank : 3) | θ value | 1.05554e-24 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.503809 (rank : 4) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TDRD7_MOUSE
|
||||||
| NC score | 0.455819 (rank : 5) | θ value | 1.9326e-10 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K1H1, Q8R181 | Gene names | Tdrd7, Pctaire2bp | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRD7_HUMAN
|
||||||
| NC score | 0.440349 (rank : 6) | θ value | 1.06045e-08 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TDRKH_MOUSE
|
||||||
| NC score | 0.413135 (rank : 7) | θ value | 4.76016e-09 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80VL1 | Gene names | Tdrkh | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.407728 (rank : 8) | θ value | 1.38499e-08 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
SND1_HUMAN
|
||||||
| NC score | 0.394993 (rank : 9) | θ value | 3.08544e-08 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7KZF4, Q13122 | Gene names | SND1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator) (EBNA2 coactivator p100). | |||||
|
SND1_MOUSE
|
||||||
| NC score | 0.391368 (rank : 10) | θ value | 8.97725e-08 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q78PY7, Q3TT46, Q922L5, Q9R0S1 | Gene names | Snd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Staphylococcal nuclease domain-containing protein 1 (p100 co- activator) (100 kDa coactivator). | |||||
|
TDRD4_HUMAN
|
||||||
| NC score | 0.389504 (rank : 11) | θ value | 7.1131e-05 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
AKAP1_HUMAN
|
||||||
| NC score | 0.274732 (rank : 12) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92667, Q13320, Q9BW14 | Gene names | AKAP1, AKAP149 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (A-kinase anchor protein 149 kDa) (AKAP 149) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein 84) (S-AKAP84). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.265599 (rank : 13) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
STK31_MOUSE
|
||||||
| NC score | 0.215161 (rank : 14) | θ value | 6.43352e-06 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99MW1 | Gene names | Stk31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1). | |||||
|
STK31_HUMAN
|
||||||
| NC score | 0.210708 (rank : 15) | θ value | 8.40245e-06 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXU1, Q9BXH8 | Gene names | STK31 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 31 (EC 2.7.11.1) (Serine/threonine- protein kinase NYD-SPK). | |||||
|
TDRD3_HUMAN
|
||||||
| NC score | 0.151390 (rank : 16) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
TDRD3_MOUSE
|
||||||
| NC score | 0.141442 (rank : 17) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91W18, Q8BZW6 | Gene names | Tdrd3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
RNF17_HUMAN
|
||||||
| NC score | 0.123552 (rank : 18) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXT8, Q9BXT7 | Gene names | RNF17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
SPF30_HUMAN
|
||||||
| NC score | 0.122032 (rank : 19) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SPF30_MOUSE
|
||||||
| NC score | 0.115763 (rank : 20) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
RNF17_MOUSE
|
||||||
| NC score | 0.112474 (rank : 21) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MV7 | Gene names | Rnf17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
SCTM1_HUMAN
|
||||||
| NC score | 0.059501 (rank : 22) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVN6, O00466 | Gene names | SECTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secreted and transmembrane protein 1 precursor (Protein K12). | |||||
|
LYPA1_MOUSE
|
||||||
| NC score | 0.057249 (rank : 23) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97823, Q3TJZ0, Q7TPX1, Q8BWM6 | Gene names | Lypla1, Apt1, Pla1a | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-protein thioesterase 1 (EC 3.1.2.-) (Lysophospholipase 1) (Lysophospholipase I) (LysoPLA I). | |||||
|
LYPA1_HUMAN
|
||||||
| NC score | 0.051638 (rank : 24) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75608, O43202, Q9UQF9 | Gene names | LYPLA1, APT1, LPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-protein thioesterase 1 (EC 3.1.2.-) (Lysophospholipase 1) (Lysophospholipase I). | |||||
|
CJ006_HUMAN
|
||||||
| NC score | 0.047493 (rank : 25) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX21, Q5W0L8, Q9NPE8 | Gene names | C10orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6. | |||||
|
FAS_HUMAN
|
||||||
| NC score | 0.043746 (rank : 26) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49327, Q13479, Q16702, Q6P4U5, Q6SS02, Q969R1, Q96C68, Q96IT0 | Gene names | FASN, FAS | |||
|
Domain Architecture |

|
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
FAS_MOUSE
|
||||||
| NC score | 0.039594 (rank : 27) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19096, Q6PB72, Q8C4Z0, Q9EQR0 | Gene names | Fasn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fatty acid synthase (EC 2.3.1.85) [Includes: [Acyl-carrier-protein] S- acetyltransferase (EC 2.3.1.38); [Acyl-carrier-protein] S- malonyltransferase (EC 2.3.1.39); 3-oxoacyl-[acyl-carrier-protein] synthase (EC 2.3.1.41); 3-oxoacyl-[acyl-carrier-protein] reductase (EC 1.1.1.100); 3-hydroxypalmitoyl-[acyl-carrier-protein] dehydratase (EC 4.2.1.61); Enoyl-[acyl-carrier-protein] reductase (EC 1.3.1.10); Oleoyl-[acyl-carrier-protein] hydrolase (EC 3.1.2.14)]. | |||||
|
U384_HUMAN
|
||||||
| NC score | 0.038210 (rank : 28) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y3C1, Q6IAI6, Q8IXL5 | Gene names | ames=CGI-117, HSPC111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0384 protein CGI-117 (HBV pre-S2 trans-regulated protein 3). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.032955 (rank : 29) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.030632 (rank : 30) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.030133 (rank : 31) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.029431 (rank : 32) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.027135 (rank : 33) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
HMGN3_MOUSE
|
||||||
| NC score | 0.026444 (rank : 34) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCB1, Q7M737, Q99JU1 | Gene names | Hmgn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group nucleosome-binding domain-containing protein 3. | |||||
|
GDPD2_MOUSE
|
||||||
| NC score | 0.025824 (rank : 35) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ESM6 | Gene names | Gdpd2, Gde3, Obdpf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycerophosphodiester phosphodiesterase domain-containing protein 2 (EC 3.1.-.-) (Glycerophosphodiester phosphodiesterase 3) (Osteoblast differentiation promoting factor). | |||||
|
GIDRP_MOUSE
|
||||||
| NC score | 0.025236 (rank : 36) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJM3, Q3UGJ8, Q5U5U8 | Gene names | Gidrp88, D19Ertd386e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth inhibition and differentiation-related protein 88 homolog. | |||||
|
TTC16_MOUSE
|
||||||
| NC score | 0.024309 (rank : 37) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C1F5 | Gene names | Ttc16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
APLP1_HUMAN
|
||||||
| NC score | 0.022894 (rank : 38) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51693, O00113, Q96A92 | Gene names | APLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
FBX38_MOUSE
|
||||||
| NC score | 0.022402 (rank : 39) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.022241 (rank : 40) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.020868 (rank : 41) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SNTB1_HUMAN
|
||||||
| NC score | 0.020356 (rank : 42) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13884, O14912, Q4KMG8 | Gene names | SNTB1, SNT2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Tax interaction protein 43) (TIP-43) (Syntrophin 2) (BSYN2). | |||||
|
ACET_HUMAN
|
||||||
| NC score | 0.019938 (rank : 43) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22966 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, testis-specific isoform precursor (EC 3.4.15.1) (EC 3.2.1.-) (ACE-T) (Dipeptidyl carboxypeptidase I) (Kininase II) [Contains: Angiotensin-converting enzyme, testis- specific isoform, soluble form]. | |||||
|
ACE_HUMAN
|
||||||
| NC score | 0.019521 (rank : 44) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12821, Q53YX9, Q59GY8 | Gene names | ACE, DCP, DCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Angiotensin-converting enzyme, somatic isoform precursor (EC 3.4.15.1) (Dipeptidyl carboxypeptidase I) (Kininase II) (CD143 antigen) [Contains: Angiotensin-converting enzyme, somatic isoform, soluble form]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.019499 (rank : 45) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.018451 (rank : 46) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
NUDT4_HUMAN
|
||||||
| NC score | 0.017278 (rank : 47) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZJ9, Q53EZ2, Q68DD7, Q9NPC5, Q9NS30, Q9NZK0, Q9NZK1 | Gene names | NUDT4, DIPP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diphosphoinositol polyphosphate phosphohydrolase 2 (EC 3.6.1.52) (DIPP-2) (Diadenosine 5',5'''-P1,P6-hexaphosphate hydrolase 2) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 4) (Nudix motif 4). | |||||
|
NUDT4_MOUSE
|
||||||
| NC score | 0.017254 (rank : 48) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2U6, Q8BXB9, Q9D3T6 | Gene names | Nudt4, Dipp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diphosphoinositol polyphosphate phosphohydrolase 2 (EC 3.6.1.52) (DIPP-2) (Diadenosine 5',5'''-P1,P6-hexaphosphate hydrolase 2) (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 4) (Nudix motif 4). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.016461 (rank : 49) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.016255 (rank : 50) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.016175 (rank : 51) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
FA8_HUMAN
|
||||||
| NC score | 0.016118 (rank : 52) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00451 | Gene names | F8, F8C | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VIII precursor (Procoagulant component) (Antihemophilic factor) (AHF) [Contains: Factor VIIIa heavy chain, 200 kDa isoform; Factor VIIIa heavy chain, 92 kDa isoform; Factor VIII B chain; Factor VIIIa light chain]. | |||||
|
SNTB1_MOUSE
|
||||||
| NC score | 0.015391 (rank : 53) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99L88, O35925 | Gene names | Sntb1, Snt2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Syntrophin 2). | |||||
|
EPMIP_HUMAN
|
||||||
| NC score | 0.015095 (rank : 54) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7L775, O94866, Q9H3L3 | Gene names | EPM2AIP1, KIAA0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.013718 (rank : 55) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.013188 (rank : 56) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
F10A5_HUMAN
|
||||||
| NC score | 0.012678 (rank : 57) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFI4 | Gene names | FAM10A5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM10A5. | |||||
|
AQR_MOUSE
|
||||||
| NC score | 0.011934 (rank : 58) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFQ3, P97871, Q3U9N1, Q3ULE8, Q80TX8 | Gene names | Aqr, Kiaa0560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intron-binding protein aquarius. | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.011912 (rank : 59) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.011730 (rank : 60) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.011606 (rank : 61) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
SPA12_MOUSE
|
||||||
| NC score | 0.011368 (rank : 62) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TMF5, Q9CQ32 | Gene names | Serpina12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serpin A12 precursor (Visceral adipose-specific serpin) (Visceral adipose tissue-derived serine protease inhibitor) (Vaspin). | |||||
|
F13A_MOUSE
|
||||||
| NC score | 0.010624 (rank : 63) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BH61, Q3TNF9, Q8BIP2 | Gene names | F13a1, F13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coagulation factor XIII A chain precursor (EC 2.3.2.13) (Coagulation factor XIIIa) (Protein-glutamine gamma-glutamyltransferase A chain) (Transglutaminase A chain). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.010568 (rank : 64) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
PAX5_HUMAN
|
||||||
| NC score | 0.010111 (rank : 65) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02548, O75933 | Gene names | PAX5 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PAX5_MOUSE
|
||||||
| NC score | 0.010103 (rank : 66) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02650 | Gene names | Pax5, Pax-5 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
CSKI2_HUMAN
|
||||||
| NC score | 0.009401 (rank : 67) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXE0, Q7LG69, Q9ULT1 | Gene names | CASKIN2, KIAA1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
PTPRD_MOUSE
|
||||||
| NC score | 0.008646 (rank : 68) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64487, Q64486, Q64488, Q64495 | Gene names | Ptprd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | 0.008298 (rank : 69) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
IRX5_MOUSE
|
||||||
| NC score | 0.007825 (rank : 70) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKQ4, Q80WV4, Q9JLL5 | Gene names | Irx5, Irxb2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.007763 (rank : 71) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
FBLN4_MOUSE
|
||||||
| NC score | 0.007656 (rank : 72) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVJ9 | Gene names | Efemp2, Fbln4, Mbp1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (Mutant p53-binding protein 1). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.007636 (rank : 73) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
FA10_MOUSE
|
||||||
| NC score | 0.007528 (rank : 74) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FBLN4_HUMAN
|
||||||
| NC score | 0.006272 (rank : 75) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.004571 (rank : 76) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
WNK4_MOUSE
|
||||||
| NC score | 0.004504 (rank : 77) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.004490 (rank : 78) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CP1A1_HUMAN
|
||||||
| NC score | 0.004441 (rank : 79) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04798 | Gene names | CYP1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1) (P450 form 6) (P450-C). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.004281 (rank : 80) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
KLK5_HUMAN
|
||||||
| NC score | 0.003437 (rank : 81) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y337, Q9HBG8 | Gene names | KLK5, SCTE | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-5 precursor (EC 3.4.21.-) (Stratum corneum tryptic enzyme) (Kallikrein-like protein 2) (KLK-L2). | |||||
|
PASK_HUMAN
|
||||||
| NC score | 0.000515 (rank : 82) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 857 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RG2, Q86XH6, Q99763, Q9UFR7 | Gene names | PASK, KIAA0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN) (hPASK). | |||||