Please be patient as the page loads
|
SPF30_HUMAN
|
||||||
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPF30_HUMAN
|
||||||
| θ value | 4.93473e-115 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SPF30_MOUSE
|
||||||
| θ value | 5.45601e-114 (rank : 2) | NC score | 0.997751 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SMN_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 3) | NC score | 0.539585 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97801, O09092 | Gene names | Smn1, Smn | |||
|
Domain Architecture |
|
|||||
| Description | Survival motor neuron protein. | |||||
|
SMN_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 4) | NC score | 0.522560 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |
|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
TDRD3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 5) | NC score | 0.420416 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91W18, Q8BZW6 | Gene names | Tdrd3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
TDRD3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 6) | NC score | 0.409881 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 7) | NC score | 0.275203 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 8) | NC score | 0.238459 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
TDRD1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.181744 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 10) | NC score | 0.161501 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.071962 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.036363 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.044003 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.042265 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.033911 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
TDRD6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.122032 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
GDF8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.030033 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14793, Q6B0H2 | Gene names | GDF8, MSTN | |||
|
Domain Architecture |
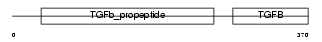
|
|||||
| Description | Growth/differentiation factor 8 precursor (GDF-8) (Myostatin). | |||||
|
GDF8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.030081 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08689 | Gene names | Gdf8, Mstn | |||
|
Domain Architecture |
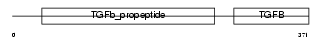
|
|||||
| Description | Growth/differentiation factor 8 precursor (GDF-8) (Myostatin). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.021213 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.025713 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.032663 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.032595 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.040047 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
SPC24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.040747 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
CING_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.021383 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
SEPT7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.016509 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16181, Q6NX50 | Gene names | SEPT7, CDC10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
CCD42_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.026875 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96M95, Q8N6Q0 | Gene names | CCDC42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 42. | |||||
|
KIF14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.012275 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
OPA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.021450 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.015342 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.018755 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
DDX55_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.007343 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
SPF30_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.93473e-115 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SPF30_MOUSE
|
||||||
| NC score | 0.997751 (rank : 2) | θ value | 5.45601e-114 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SMN_MOUSE
|
||||||
| NC score | 0.539585 (rank : 3) | θ value | 5.62301e-10 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97801, O09092 | Gene names | Smn1, Smn | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein. | |||||
|
SMN_HUMAN
|
||||||
| NC score | 0.522560 (rank : 4) | θ value | 1.80886e-08 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16637, Q13119, Q96J51 | Gene names | SMN1, SMN, SMNT | |||
|
Domain Architecture |

|
|||||
| Description | Survival motor neuron protein (Component of gems 1) (Gemin-1). | |||||
|
TDRD3_MOUSE
|
||||||
| NC score | 0.420416 (rank : 5) | θ value | 1.17247e-07 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91W18, Q8BZW6 | Gene names | Tdrd3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
TDRD3_HUMAN
|
||||||
| NC score | 0.409881 (rank : 6) | θ value | 1.99992e-07 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7E2, Q6P992 | Gene names | TDRD3 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 3. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.275203 (rank : 7) | θ value | 4.1701e-05 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.238459 (rank : 8) | θ value | 0.00298849 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
TDRD1_MOUSE
|
||||||
| NC score | 0.181744 (rank : 9) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99MV1 | Gene names | Tdrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD1_HUMAN
|
||||||
| NC score | 0.161501 (rank : 10) | θ value | 0.0736092 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BXT4, Q9H7B3 | Gene names | TDRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 1. | |||||
|
TDRD6_MOUSE
|
||||||
| NC score | 0.122032 (rank : 11) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P61407 | Gene names | Tdrd6 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 6. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.071962 (rank : 12) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.044003 (rank : 13) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.042265 (rank : 14) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SPC24_HUMAN
|
||||||
| NC score | 0.040747 (rank : 15) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.040047 (rank : 16) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.036363 (rank : 17) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.033911 (rank : 18) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.032663 (rank : 19) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.032595 (rank : 20) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
GDF8_MOUSE
|
||||||
| NC score | 0.030081 (rank : 21) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08689 | Gene names | Gdf8, Mstn | |||
|
Domain Architecture |

|
|||||
| Description | Growth/differentiation factor 8 precursor (GDF-8) (Myostatin). | |||||
|
GDF8_HUMAN
|
||||||
| NC score | 0.030033 (rank : 22) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14793, Q6B0H2 | Gene names | GDF8, MSTN | |||
|
Domain Architecture |

|
|||||
| Description | Growth/differentiation factor 8 precursor (GDF-8) (Myostatin). | |||||
|
CCD42_HUMAN
|
||||||
| NC score | 0.026875 (rank : 23) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96M95, Q8N6Q0 | Gene names | CCDC42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 42. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.025713 (rank : 24) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
OPA1_HUMAN
|
||||||
| NC score | 0.021450 (rank : 25) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60313 | Gene names | OPA1, KIAA0567 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-like 120 kDa protein, mitochondrial precursor (Optic atrophy protein 1) [Contains: Dynamin-like 120 kDa protein, form S1]. | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.021383 (rank : 26) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.021213 (rank : 27) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.018755 (rank : 28) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
SEPT7_HUMAN
|
||||||
| NC score | 0.016509 (rank : 29) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16181, Q6NX50 | Gene names | SEPT7, CDC10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.015342 (rank : 30) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
KIF14_HUMAN
|
||||||
| NC score | 0.012275 (rank : 31) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15058 | Gene names | KIF14, KIAA0042 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF14. | |||||
|
DDX55_HUMAN
|
||||||
| NC score | 0.007343 (rank : 32) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHQ9, Q658L6, Q9HCH7 | Gene names | DDX55, KIAA1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||