Please be patient as the page loads
|
ZGPAT_MOUSE
|
||||||
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975006 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
TFP11_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 3) | NC score | 0.383953 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 4) | NC score | 0.383948 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 5) | NC score | 0.230145 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
SPF30_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.238459 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |
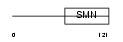
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SPF30_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 7) | NC score | 0.240724 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |
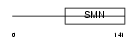
|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
GPTC2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 8) | NC score | 0.235738 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.231663 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.088049 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 11) | NC score | 0.128713 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
GPTC3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.175257 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
BAT4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.162892 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |
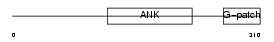
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
RBM5_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.195592 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.195349 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
BAT4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.153659 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
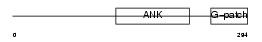
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
STMN4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.067147 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |
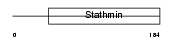
|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
STMN4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.067131 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.110458 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.059706 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.052535 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
NU155_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.079483 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99P88 | Gene names | Nup155 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup155 (Nucleoporin Nup155) (155 kDa nucleoporin). | |||||
|
GPTC3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.153152 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.051093 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
K0082_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.126951 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.098222 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.041353 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
SF04_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.155986 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.151866 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.058335 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.050970 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NU155_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.071287 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75694, Q9UBE9, Q9UFL5 | Gene names | NUP155, KIAA0791 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup155 (Nucleoporin Nup155) (155 kDa nucleoporin). | |||||
|
SF04_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.150866 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.050917 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NKRF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.151083 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
PINX1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.168648 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.041494 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UACA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.042260 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
VPK3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.124913 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.081045 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.051852 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NKRF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.150286 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.038144 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.041006 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
VPK7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.128140 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.049787 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.052235 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.093587 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.018344 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.052841 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.076171 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.056111 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
K1C14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.027418 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |
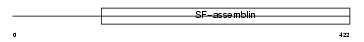
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.028752 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.091769 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.052824 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
SFR14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.145167 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
VPK10_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.126106 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
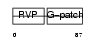
|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.126092 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.125928 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.126336 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.126691 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.136268 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.051384 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.011457 (rank : 112) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.038132 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
VPK17_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.123664 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.120992 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.134199 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
CING_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.048410 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.069026 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.067144 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
K0082_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.121622 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.080985 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.049658 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.037679 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.045077 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CKAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.046907 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.012132 (rank : 111) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.042995 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.017948 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
K1C14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.026366 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |
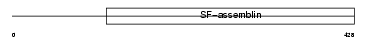
|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.079080 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
PINX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.150631 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
SYAM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.016973 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JTZ9, Q8N198, Q96D02, Q9ULF0 | Gene names | AARSL, AARS2, KIAA1270 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable alanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.7) (Alanine--tRNA ligase) (AlaRS) (Alanyl-tRNA synthetase like). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.036762 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TDRD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.039461 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
ZNFX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.014905 (rank : 110) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
ANR26_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.036675 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.043286 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.053561 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
K1C19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.016871 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08727, Q5XG83, Q6NW33, Q7L5M9, Q96A53, Q96FV1, Q9BYF9, Q9P1Y4 | Gene names | KRT19 | |||
|
Domain Architecture |
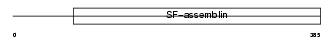
|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.040852 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
SPC24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.049942 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
VPK11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.114292 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.020450 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.007807 (rank : 113) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CEP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.030545 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.051229 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.042936 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.043416 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.034373 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
NUCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.029916 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
NUPL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.043638 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.039473 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.031754 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.032354 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.056759 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.074297 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.050133 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RBM6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.059565 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
VPK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.095718 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.095752 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.975006 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
TFP11_HUMAN
|
||||||
| NC score | 0.383953 (rank : 3) | θ value | 1.53129e-07 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBB9, O95908, Q5H8V8, Q9UGV7, Q9Y2Q8 | Gene names | TFIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11. | |||||
|
TFP11_MOUSE
|
||||||
| NC score | 0.383948 (rank : 4) | θ value | 1.53129e-07 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ERA6, Q8VD06 | Gene names | Tfip11, Tip39 | |||
|
Domain Architecture |

|
|||||
| Description | Tuftelin-interacting protein 11 (Tuftelin-interacting protein 39). | |||||
|
SPF30_MOUSE
|
||||||
| NC score | 0.240724 (rank : 5) | θ value | 0.00298849 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BGT7, Q4QQL0 | Gene names | Smndc1, Smnr, Spf30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
SPF30_HUMAN
|
||||||
| NC score | 0.238459 (rank : 6) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75940 | Gene names | SMNDC1, SMNR, SPF30 | |||
|
Domain Architecture |

|
|||||
| Description | Survival of motor neuron-related-splicing factor 30 (SMN-related protein) (30 kDa splicing factor SMNrp) (Survival motor neuron domain- containing protein 1). | |||||
|
GPTC2_MOUSE
|
||||||
| NC score | 0.235738 (rank : 7) | θ value | 0.0113563 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TQC7, Q8BNJ9, Q8BPM1, Q8CDH9 | Gene names | Gpatc2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
GPTC2_HUMAN
|
||||||
| NC score | 0.231663 (rank : 8) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NW75, Q86YE7 | Gene names | GPATC2 | |||
|
Domain Architecture |

|
|||||
| Description | G patch domain-containing protein 2. | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.230145 (rank : 9) | θ value | 0.00228821 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
RBM5_HUMAN
|
||||||
| NC score | 0.195592 (rank : 10) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P52756, Q93021, Q9BU14, Q9UKY8, Q9UL24 | Gene names | RBM5 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15) (Protein G15) (NY-REN-9 antigen). | |||||
|
RBM5_MOUSE
|
||||||
| NC score | 0.195349 (rank : 11) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91YE7, Q3UZ19, Q99KV9 | Gene names | Rbm5, Luca15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 5 (RNA-binding motif protein 5) (Tumor suppressor LUCA15). | |||||
|
GPTC3_MOUSE
|
||||||
| NC score | 0.175257 (rank : 12) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BIY1 | Gene names | Gpatc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
PINX1_MOUSE
|
||||||
| NC score | 0.168648 (rank : 13) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9CZX5, Q91WZ9, Q9D0C2 | Gene names | Pinx1, Lpts | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (LPTS1) (67-11-3 protein). | |||||
|
BAT4_MOUSE
|
||||||
| NC score | 0.162892 (rank : 14) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61858, Q9R065 | Gene names | Bat4, Bat-4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.155986 (rank : 15) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.153659 (rank : 16) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
GPTC3_HUMAN
|
||||||
| NC score | 0.153152 (rank : 17) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96I76, Q5JYH2, Q8NDJ2, Q9H9Z3 | Gene names | GPATC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 3. | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.151866 (rank : 18) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
NKRF_MOUSE
|
||||||
| NC score | 0.151083 (rank : 19) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BY02, Q8BJU5, Q9CRL2 | Gene names | Nkrf, Nrf | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF). | |||||
|
SF04_MOUSE
|
||||||
| NC score | 0.150866 (rank : 20) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CH02, Q8R094 | Gene names | Sf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4. | |||||
|
PINX1_HUMAN
|
||||||
| NC score | 0.150631 (rank : 21) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96BK5, Q7Z7J8, Q96QD7, Q9HBU7, Q9NWW2 | Gene names | PINX1, LPTS | |||
|
Domain Architecture |

|
|||||
| Description | Pin2-interacting protein X1 (TRF1-interacting protein 1) (Liver- related putative tumor suppressor) (67-11-3 protein). | |||||
|
NKRF_HUMAN
|
||||||
| NC score | 0.150286 (rank : 22) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15226, Q9UJ91 | Gene names | NKRF, ITBA4, NRF | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B-repressing factor (NFKB-repressing factor) (Transcription factor NRF) (ITBA4 protein). | |||||
|
SFR14_HUMAN
|
||||||
| NC score | 0.145167 (rank : 23) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8IX01, O15071, O60369, Q5JPH7, Q8WUF7 | Gene names | SFRS14, KIAA0365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.136268 (rank : 24) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.134199 (rank : 25) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.128713 (rank : 26) | θ value | 0.0252991 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
VPK7_HUMAN
|
||||||
| NC score | 0.128140 (rank : 27) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63123 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K18 Pro protein) (HERV-K110 Pro protein) (HERV-K(C1a) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
K0082_HUMAN
|
||||||
| NC score | 0.126951 (rank : 28) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8N1G2, Q14670, Q96FJ9 | Gene names | KIAA0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
VPK9_HUMAN
|
||||||
| NC score | 0.126691 (rank : 29) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63124 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K104 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK5_HUMAN
|
||||||
| NC score | 0.126336 (rank : 30) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63121 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K113 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
VPK10_HUMAN
|
||||||
| NC score | 0.126106 (rank : 31) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10265, Q9UKH6 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-K_5q33.3 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K10 Pro protein) (HERV-K107 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK12_HUMAN
|
||||||
| NC score | 0.126092 (rank : 32) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63131, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K102 Pro protein) (HERV-K(III) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK1_HUMAN
|
||||||
| NC score | 0.125928 (rank : 33) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63119 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
VPK3_HUMAN
|
||||||
| NC score | 0.124913 (rank : 34) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63120 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K(C19) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK17_HUMAN
|
||||||
| NC score | 0.123664 (rank : 35) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63125 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Pro protein (EC 3.4.23.-) (Protease) (Proteinase) (PR). | |||||
|
K0082_MOUSE
|
||||||
| NC score | 0.121622 (rank : 36) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBC3, Q3U3G5, Q3U7Y9, Q6A0D5, Q8C7V0 | Gene names | Kiaa0082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0082. | |||||
|
VPK2_HUMAN
|
||||||
| NC score | 0.120992 (rank : 37) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6I0, Q9UKH5 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K(HML-2.HOM) Pro protein) (HERV-K108 Pro protein) (HERV-K(C7) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK11_HUMAN
|
||||||
| NC score | 0.114292 (rank : 38) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P63129, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Pro protein (EC 3.4.23.-) (HERV- K101 envelope protein) (Protease) (Proteinase) (PR). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.110458 (rank : 39) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.098222 (rank : 40) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
VPK6_HUMAN
|
||||||
| NC score | 0.095752 (rank : 41) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63122 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K115 Pro protein) (Protease) (Proteinase) (PR). | |||||
|
VPK4_HUMAN
|
||||||
| NC score | 0.095718 (rank : 42) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63127, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Pro protein (EC 3.4.23.-) (HERV-K109 Pro protein) (HERV-K(C6) Pro protein) (Protease) (Proteinase) (PR). | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.093587 (rank : 43) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.091769 (rank : 44) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.088049 (rank : 45) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.081045 (rank : 46) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.080985 (rank : 47) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
NU155_MOUSE
|
||||||
| NC score | 0.079483 (rank : 48) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99P88 | Gene names | Nup155 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup155 (Nucleoporin Nup155) (155 kDa nucleoporin). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.079080 (rank : 49) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.076171 (rank : 50) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.074297 (rank : 51) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
NU155_HUMAN
|
||||||
| NC score | 0.071287 (rank : 52) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75694, Q9UBE9, Q9UFL5 | Gene names | NUP155, KIAA0791 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup155 (Nucleoporin Nup155) (155 kDa nucleoporin). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.069026 (rank : 53) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
STMN4_HUMAN
|
||||||
| NC score | 0.067147 (rank : 54) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H169 | Gene names | STMN4 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.067144 (rank : 55) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
STMN4_MOUSE
|
||||||
| NC score | 0.067131 (rank : 56) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P63042, O35414, O35415, O35416 | Gene names | Stmn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stathmin-4 (Stathmin-like protein B3) (RB3). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.059706 (rank : 57) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RBM6_HUMAN
|
||||||
| NC score | 0.059565 (rank : 58) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78332, O60549, O75524 | Gene names | RBM6, DEF3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 6 (RNA-binding motif protein 6) (RNA-binding protein DEF-3) (Lung cancer antigen NY-LU-12) (Protein G16). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.058335 (rank : 59) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.056759 (rank : 60) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.056111 (rank : 61) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.053561 (rank : 62) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.052841 (rank : 63) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.052824 (rank : 64) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.052535 (rank : 65) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.052235 (rank : 66) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.051852 (rank : 67) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.051384 (rank : 68) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.051229 (rank : 69) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.051093 (rank : 70) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.050970 (rank : 71) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.050917 (rank : 72) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.050133 (rank : 73) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
SPC24_HUMAN
|
||||||
| NC score | 0.049942 (rank : 74) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NBT2 | Gene names | SPBC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc24 (hSpc24). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.049787 (rank : 75) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.049658 (rank : 76) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.048410 (rank : 77) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CKAP4_MOUSE
|
||||||
| NC score | 0.046907 (rank : 78) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.045077 (rank : 79) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
NUPL2_HUMAN
|
||||||
| NC score | 0.043638 (rank : 80) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.043416 (rank : 81) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.043286 (rank : 82) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.042995 (rank : 83) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.042936 (rank : 84) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.042260 (rank : 85) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.041494 (rank : 86) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.041353 (rank : 87) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.041006 (rank : 88) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.040852 (rank : 89) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.039473 (rank : 90) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
TDRD4_HUMAN
|
||||||
| NC score | 0.039461 (rank : 91) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUY9 | Gene names | TDRD4 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 4. | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.038144 (rank : 92) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.038132 (rank : 93) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.037679 (rank : 94) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.036762 (rank : 95) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ANR26_MOUSE
|
||||||
| NC score | 0.036675 (rank : 96) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1554 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q811D2, Q69ZS2, Q9CS61 | Gene names | Ankrd26, Kiaa1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.034373 (rank : 97) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.032354 (rank : 98) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.031754 (rank : 99) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
CEP35_HUMAN
|
||||||
| NC score | 0.030545 (rank : 100) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VT06, O75068, Q8TDK3, Q8WY20 | Gene names | CEP350, CAP350, GM133 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein 350 (Centrosome-associated protein of 350 kDa). | |||||
|
NUCB1_HUMAN
|
||||||
| NC score | 0.029916 (rank : 101) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q02818, Q15838, Q7Z4J7, Q9BUR1 | Gene names | NUCB1, NUC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-1 precursor (CALNUC). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.028752 (rank : 102) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
K1C14_HUMAN
|
||||||
| NC score | 0.027418 (rank : 103) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02533, Q14715, Q53XY3, Q9BUE3, Q9UBN2, Q9UBN3, Q9UCY4 | Gene names | KRT14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
K1C14_MOUSE
|
||||||
| NC score | 0.026366 (rank : 104) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61781, Q91VQ4 | Gene names | Krt14, Krt1-14 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 14 (Cytokeratin-14) (CK-14) (Keratin-14) (K14). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.020450 (rank : 105) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.018344 (rank : 106) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.017948 (rank : 107) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SYAM_HUMAN
|
||||||
| NC score | 0.016973 (rank : 108) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5JTZ9, Q8N198, Q96D02, Q9ULF0 | Gene names | AARSL, AARS2, KIAA1270 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable alanyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.7) (Alanine--tRNA ligase) (AlaRS) (Alanyl-tRNA synthetase like). | |||||
|
K1C19_HUMAN
|
||||||
| NC score | 0.016871 (rank : 109) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08727, Q5XG83, Q6NW33, Q7L5M9, Q96A53, Q96FV1, Q9BYF9, Q9P1Y4 | Gene names | KRT19 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 19 (Cytokeratin-19) (CK-19) (Keratin-19) (K19). | |||||
|
ZNFX1_HUMAN
|
||||||
| NC score | 0.014905 (rank : 110) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2E3, Q9BQM7, Q9BQM8, Q9H8C1, Q9H9S2, Q9NUM1, Q9NWW1 | Gene names | ZNFX1, KIAA1404 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NFX1-type zinc finger-containing protein 1. | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.012132 (rank : 111) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.011457 (rank : 112) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.007807 (rank : 113) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||