Please be patient as the page loads
|
PAR12_MOUSE
|
||||||
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PAR12_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995962 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZCC2_HUMAN
|
||||||
| θ value | 9.41505e-74 (rank : 3) | NC score | 0.892689 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | 5.55816e-50 (rank : 4) | NC score | 0.851758 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | 9.48078e-50 (rank : 5) | NC score | 0.849420 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
PAR15_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 6) | NC score | 0.776847 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q460N3, Q8N1K3 | Gene names | PARP15, BAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 15 (EC 2.4.2.30) (PARP-15) (B-aggressive lymphoma protein 3). | |||||
|
PAR14_MOUSE
|
||||||
| θ value | 8.91499e-32 (rank : 7) | NC score | 0.689306 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PAR14_HUMAN
|
||||||
| θ value | 6.38894e-30 (rank : 8) | NC score | 0.692945 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q460N5, Q460N4, Q8J027, Q9H9X9, Q9NV60, Q9ULF2 | Gene names | PARP14, BAL2, KIAA1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 9) | NC score | 0.659517 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 10) | NC score | 0.153663 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
PARP9_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 11) | NC score | 0.538033 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
PARP9_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 12) | NC score | 0.512108 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |
|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 13) | NC score | 0.130458 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 14) | NC score | 0.077139 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.128763 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.103159 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.058427 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.119381 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.129935 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
NUPL2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.102495 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.116914 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.115461 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.116530 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.115634 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
NUPL2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.094507 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.114074 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.123420 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.105840 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.106837 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.088002 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.102564 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
ZN185_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.035862 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62394 | Gene names | Znf185, Zfp185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein Zfp185) (P1-A). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.059760 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.059760 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.030862 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PO2F2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.010308 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00196, Q00197, Q00198, Q00199, Q00200, Q00201, Q05882, Q61995, Q61996, Q64245 | Gene names | Pou2f2, Oct2, Otf2 | |||
|
Domain Architecture |
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.015339 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.080947 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.027167 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.008633 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.011837 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
EMIL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.010529 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.001330 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
ZSWM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.029451 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
ZSWM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.026325 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.008158 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.015341 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.016804 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.005932 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
PARP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.015268 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2NL67, Q9H7C5, Q9H9X6, Q9HAF3, Q9NPS6, Q9UFG4 | Gene names | PARP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PARP6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.015265 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P6P7, Q8BVW1 | Gene names | Parp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PO2F2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.007907 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |
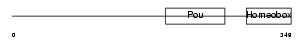
|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.009235 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
STAR7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.017789 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |
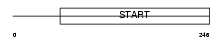
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
ZGPAT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.035225 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.087978 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
STAR7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.016747 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |
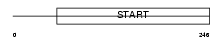
|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.003811 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
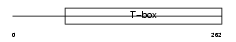
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.068372 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
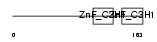
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
ZGPAT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.032354 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
H2AW_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.057357 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AW_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056432 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.073086 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.072791 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
LRP16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.162944 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |
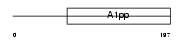
|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.156780 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |
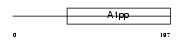
|
|||||
| Description | Protein LRP16. | |||||
|
PARP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.086771 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.076780 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.066586 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.995962 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
ZCC2_HUMAN
|
||||||
| NC score | 0.892689 (rank : 3) | θ value | 9.41505e-74 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.851758 (rank : 4) | θ value | 5.55816e-50 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.849420 (rank : 5) | θ value | 9.48078e-50 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
PAR15_HUMAN
|
||||||
| NC score | 0.776847 (rank : 6) | θ value | 5.22648e-32 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q460N3, Q8N1K3 | Gene names | PARP15, BAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 15 (EC 2.4.2.30) (PARP-15) (B-aggressive lymphoma protein 3). | |||||
|
PAR14_HUMAN
|
||||||
| NC score | 0.692945 (rank : 7) | θ value | 6.38894e-30 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q460N5, Q460N4, Q8J027, Q9H9X9, Q9NV60, Q9ULF2 | Gene names | PARP14, BAL2, KIAA1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (B aggressive lymphoma protein 2). | |||||
|
PAR14_MOUSE
|
||||||
| NC score | 0.689306 (rank : 8) | θ value | 8.91499e-32 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2EMV9, Q571C6, Q8BN35, Q8VDT6 | Gene names | Parp14, Kiaa1268 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 14 (EC 2.4.2.30) (PARP-14) (Collaborator of STAT6) (CoaSt6). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.659517 (rank : 9) | θ value | 5.07402e-19 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
PARP9_HUMAN
|
||||||
| NC score | 0.538033 (rank : 10) | θ value | 8.11959e-09 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXQ6, Q8TCP3, Q9BZL8, Q9BZL9 | Gene names | PARP9, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein). | |||||
|
PARP9_MOUSE
|
||||||
| NC score | 0.512108 (rank : 11) | θ value | 1.06045e-08 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CAS9, Q99LF9 | Gene names | Parp9, Bal | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 9 (EC 2.4.2.30) (PARP-9) (B aggressive lymphoma protein homolog). | |||||
|
LRP16_HUMAN
|
||||||
| NC score | 0.162944 (rank : 12) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ69, Q9UH96 | Gene names | LRP16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
LRP16_MOUSE
|
||||||
| NC score | 0.156780 (rank : 13) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922B1 | Gene names | Lrp16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein LRP16. | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.153663 (rank : 14) | θ value | 9.59137e-10 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.130458 (rank : 15) | θ value | 1.80886e-08 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.129935 (rank : 16) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.128763 (rank : 17) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.123420 (rank : 18) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.119381 (rank : 19) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.116914 (rank : 20) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.116530 (rank : 21) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.115634 (rank : 22) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.115461 (rank : 23) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.114074 (rank : 24) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.106837 (rank : 25) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.105840 (rank : 26) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.103159 (rank : 27) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.102564 (rank : 28) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
NUPL2_HUMAN
|
||||||
| NC score | 0.102495 (rank : 29) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15504, Q49AE7, Q9BS49 | Gene names | NUPL2, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1) (hCG1) (NUP42 homolog). | |||||
|
NUPL2_MOUSE
|
||||||
| NC score | 0.094507 (rank : 30) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIC2, Q8BJX3, Q8BVP7, Q924S0 | Gene names | Nupl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin-like 2 (NLP-1). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.088002 (rank : 31) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.087978 (rank : 32) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
PARP3_HUMAN
|
||||||
| NC score | 0.086771 (rank : 33) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y6F1, Q8NER9, Q96CG2, Q9UG81 | Gene names | PARP3, ADPRT3, ADPRTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 3 (EC 2.4.2.30) (PARP-3) (NAD(+) ADP- ribosyltransferase 3) (Poly[ADP-ribose] synthetase 3) (pADPRT-3) (hPARP-3) (IRT1). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.080947 (rank : 34) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.077139 (rank : 35) | θ value | 0.00665767 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.076780 (rank : 36) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
H2AY_HUMAN
|
||||||
| NC score | 0.073086 (rank : 37) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75367, O75377, Q503A8, Q7Z5E3, Q96D41, Q9H8P3, Q9UP96 | Gene names | H2AFY, MACROH2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) (Medulloblastoma antigen MU-MB-50.205). | |||||
|
H2AY_MOUSE
|
||||||
| NC score | 0.072791 (rank : 38) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZQ8, Q91VZ2, Q9QZQ7 | Gene names | H2afy | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y). | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.068372 (rank : 39) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.066586 (rank : 40) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.059760 (rank : 41) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.059760 (rank : 42) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.058427 (rank : 43) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
H2AW_HUMAN
|
||||||
| NC score | 0.057357 (rank : 44) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P0M6, Q5SQT2 | Gene names | H2AFY2, MACROH2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
H2AW_MOUSE
|
||||||
| NC score | 0.056432 (rank : 45) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCK0, Q3TNT4, Q925I6 | Gene names | H2afy2, H2afy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2). | |||||
|
ZN185_MOUSE
|
||||||
| NC score | 0.035862 (rank : 46) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62394 | Gene names | Znf185, Zfp185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein Zfp185) (P1-A). | |||||
|
ZGPAT_HUMAN
|
||||||
| NC score | 0.035225 (rank : 47) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8N5A5, Q5JWI9, Q8NC55, Q96JI0, Q96JU4, Q9H401 | Gene names | ZGPAT, KIAA1847, ZC3H9, ZC3HDC9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein (Zinc finger CCCH domain-containing protein 9). | |||||
|
ZGPAT_MOUSE
|
||||||
| NC score | 0.032354 (rank : 48) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VDM1, Q8BWW2 | Gene names | Zgpat | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type with G patch domain protein. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.030862 (rank : 49) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
ZSWM2_HUMAN
|
||||||
| NC score | 0.029451 (rank : 50) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.027167 (rank : 51) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
ZSWM2_MOUSE
|
||||||
| NC score | 0.026325 (rank : 52) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D9X6 | Gene names | Zswim2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
STAR7_HUMAN
|
||||||
| NC score | 0.017789 (rank : 53) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQZ5, Q6GU43, Q969M6 | Gene names | STARD7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7) (Protein GTT1). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.016804 (rank : 54) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
STAR7_MOUSE
|
||||||
| NC score | 0.016747 (rank : 55) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R1R3 | Gene names | Stard7 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 7 (StARD7) (START domain- containing protein 7). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.015341 (rank : 56) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.015339 (rank : 57) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
PARP6_HUMAN
|
||||||
| NC score | 0.015268 (rank : 58) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2NL67, Q9H7C5, Q9H9X6, Q9HAF3, Q9NPS6, Q9UFG4 | Gene names | PARP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
PARP6_MOUSE
|
||||||
| NC score | 0.015265 (rank : 59) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P6P7, Q8BVW1 | Gene names | Parp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 6 (EC 2.4.2.30) (PARP-6). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.011837 (rank : 60) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
EMIL1_HUMAN
|
||||||
| NC score | 0.010529 (rank : 61) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
PO2F2_MOUSE
|
||||||
| NC score | 0.010308 (rank : 62) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00196, Q00197, Q00198, Q00199, Q00200, Q00201, Q05882, Q61995, Q61996, Q64245 | Gene names | Pou2f2, Oct2, Otf2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.009235 (rank : 63) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.008633 (rank : 64) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.008158 (rank : 65) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PO2F2_HUMAN
|
||||||
| NC score | 0.007907 (rank : 66) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09086, Q16648, Q9BRS4 | Gene names | POU2F2, OCT2, OTF2 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 2 (Octamer-binding transcription factor 2) (Oct-2) (OTF-2) (Lymphoid-restricted immunoglobulin octamer-binding protein NF-A2). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.005932 (rank : 67) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.003811 (rank : 68) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.001330 (rank : 69) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||