Please be patient as the page loads
|
CPSF4_HUMAN
|
||||||
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CPSF4_HUMAN
|
||||||
| θ value | 1.51632e-156 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 7.87842e-113 (rank : 2) | NC score | 0.986402 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
ZC3H3_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 3) | NC score | 0.641687 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 4) | NC score | 0.638360 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 5) | NC score | 0.515968 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 6) | NC score | 0.509475 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 7) | NC score | 0.569499 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 8) | NC score | 0.484192 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 9) | NC score | 0.473821 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 10) | NC score | 0.572046 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H5_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.285139 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC11A_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 12) | NC score | 0.304560 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.411502 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.293851 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
ZC3H5_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.286738 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 16) | NC score | 0.407979 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 17) | NC score | 0.378982 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 18) | NC score | 0.380564 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.326555 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.072259 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
PAR12_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.128763 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.297212 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
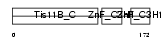
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.296009 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
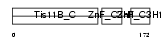
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
MBN3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 24) | NC score | 0.227484 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
TISD_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.284695 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |
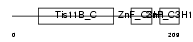
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
TISD_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.295592 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
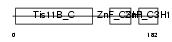
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
MBNL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.204676 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.205948 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
Z3H7A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.086236 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
MKRN3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.268602 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.262728 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
PAR12_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.131724 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
MBN3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.230331 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
R113B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.115485 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
RBM22_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.131095 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.131095 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
TTP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.289328 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |
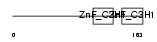
|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
CNBP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.101751 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.099704 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.052182 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.045233 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.050151 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TTP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.288998 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ZCH12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.034670 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PEW1, Q6PID5, Q8N1C1 | Gene names | ZCCHC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 12. | |||||
|
R113A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.106005 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.066773 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DUS3L_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.063947 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XI1, Q3TZQ1, Q7TT12 | Gene names | Dus3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.049183 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
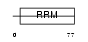
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.049136 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
ZCH13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.092974 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
ZCHC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.081825 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.057157 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
ECT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.013818 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.015207 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
TRM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.077025 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.034735 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.064902 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
DUS3L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.081472 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.022200 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
ERO1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.016484 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YB8, Q8IZ11, Q9NR62 | Gene names | ERO1LB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
ERO1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.016440 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
GRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.008156 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
NELL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.001832 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.021737 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.021299 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
TRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.052243 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TX08, Q3TBY4 | Gene names | Trmt1, D8Ertd812e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.036209 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZN615_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | -0.003211 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8J6 | Gene names | ZNF615 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 615. | |||||
|
PARPT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.051947 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PARPT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052068 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.51632e-156 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.986402 (rank : 2) | θ value | 7.87842e-113 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
ZC3H3_MOUSE
|
||||||
| NC score | 0.641687 (rank : 3) | θ value | 2.06002e-20 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CHP0, Q80X78 | Gene names | Zc3h3, Kiaa0150, Zc3hdc3, Zfp623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.638360 (rank : 4) | θ value | 1.47631e-18 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.572046 (rank : 5) | θ value | 1.17247e-07 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.569499 (rank : 6) | θ value | 6.87365e-08 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.515968 (rank : 7) | θ value | 6.21693e-09 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.509475 (rank : 8) | θ value | 5.26297e-08 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.484192 (rank : 9) | θ value | 8.97725e-08 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.473821 (rank : 10) | θ value | 8.97725e-08 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.411502 (rank : 11) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.407979 (rank : 12) | θ value | 0.000121331 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN1_MOUSE
|
||||||
| NC score | 0.380564 (rank : 13) | θ value | 0.000461057 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QXP6 | Gene names | Mkrn1 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1. | |||||
|
MKRN1_HUMAN
|
||||||
| NC score | 0.378982 (rank : 14) | θ value | 0.000270298 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UHC7, Q6GSF1, Q9H0G0, Q9UEZ7 | Gene names | MKRN1, RNF61 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-1 (RING finger protein 61). | |||||
|
MKRN4_HUMAN
|
||||||
| NC score | 0.326555 (rank : 15) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13434 | Gene names | MKRN4, RNF64, ZNF127L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Makorin-4 (Zinc finger protein 127-Xp) (ZNF127-Xp) (RING finger protein 64). | |||||
|
ZC11A_HUMAN
|
||||||
| NC score | 0.304560 (rank : 16) | θ value | 5.44631e-05 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75152, Q6AHY4, Q6AHY9, Q6AW79, Q6AWA1, Q6PJK4, Q86XZ7 | Gene names | ZC3H11A, KIAA0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.297212 (rank : 17) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.296009 (rank : 18) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.295592 (rank : 19) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.293851 (rank : 20) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
TTP_HUMAN
|
||||||
| NC score | 0.289328 (rank : 21) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P26651 | Gene names | ZFP36, G0S24, TIS11A, TTP | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36 homolog) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (G0/G1 switch regulatory protein 24). | |||||
|
TTP_MOUSE
|
||||||
| NC score | 0.288998 (rank : 22) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22893, P11520 | Gene names | Zfp36, Tis11, Tis11a | |||
|
Domain Architecture |

|
|||||
| Description | Tristetraproline (TTP) (Zinc finger protein 36) (Zfp-36) (TIS11A protein) (TIS11) (Growth factor-inducible nuclear protein NUP475) (TPA-induced sequence 11). | |||||
|
ZC3H5_MOUSE
|
||||||
| NC score | 0.286738 (rank : 23) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BL48, Q8BVI6, Q99JZ8, Q99LL9 | Gene names | Zc3h5, Kiaa1753, Zc3hdc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
ZC3H5_HUMAN
|
||||||
| NC score | 0.285139 (rank : 24) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9C0B0 | Gene names | ZC3H5, KIAA1753, ZC3HDC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 5. | |||||
|
TISD_HUMAN
|
||||||
| NC score | 0.284695 (rank : 25) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P47974, Q9BSJ3 | Gene names | ZFP36L2, BRF2, ERF2, TIS11D | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein) (EGF-response factor 2) (ERF-2). | |||||
|
MKRN3_MOUSE
|
||||||
| NC score | 0.268602 (rank : 26) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60764, Q7TQE4 | Gene names | Mkrn3, Zfp127, Znf127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.262728 (rank : 27) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
MBN3_HUMAN
|
||||||
| NC score | 0.230331 (rank : 28) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NUK0, Q5JXN8, Q5JXN9, Q5JXP4, Q6UDQ1, Q8TAD9, Q8TAF4, Q9H0Z7, Q9UF37 | Gene names | MBNL3, CHCR, MBLX39, MBXL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein HCHCR). | |||||
|
MBN3_MOUSE
|
||||||
| NC score | 0.227484 (rank : 29) | θ value | 0.0563607 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R003 | Gene names | Mbnl3, Chcr, Mbxl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like X-linked protein (Muscleblind-like protein 3) (Cys3His CCG1-required protein) (Protein MCHCR). | |||||
|
MBNL_MOUSE
|
||||||
| NC score | 0.205948 (rank : 30) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
MBNL_HUMAN
|
||||||
| NC score | 0.204676 (rank : 31) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NR56, O43311, O43797 | Gene names | MBNL1, EXP, KIAA0428, MBNL | |||
|
Domain Architecture |

|
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
PAR12_HUMAN
|
||||||
| NC score | 0.131724 (rank : 32) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H0J9, Q9H610, Q9NP36, Q9NTI3 | Gene names | PARP12, ZC3HDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
RBM22_HUMAN
|
||||||
| NC score | 0.131095 (rank : 33) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NW64, O95607 | Gene names | RBM22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
RBM22_MOUSE
|
||||||
| NC score | 0.131095 (rank : 34) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BHS3, Q3TJB0, Q9CXA0 | Gene names | Rbm22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor RBM22 (RNA-binding motif protein 22). | |||||
|
PAR12_MOUSE
|
||||||
| NC score | 0.128763 (rank : 35) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZ20, Q80VL6, Q8K333, Q8R3U2 | Gene names | Parp12, Zc3hdc1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 12 (EC 2.4.2.30) (PARP-12) (Zinc finger CCCH domain-containing protein 1). | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.115485 (rank : 36) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.106005 (rank : 37) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
CNBP_HUMAN
|
||||||
| NC score | 0.101751 (rank : 38) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| NC score | 0.099704 (rank : 39) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCH13_HUMAN
|
||||||
| NC score | 0.092974 (rank : 40) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
Z3H7A_HUMAN
|
||||||
| NC score | 0.086236 (rank : 41) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IWR0, Q9NPE9 | Gene names | ZC3H7A, ZC3H7, ZC3HDC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7A. | |||||
|
ZCHC3_HUMAN
|
||||||
| NC score | 0.081825 (rank : 42) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NUD5, Q6NT79 | Gene names | ZCCHC3, C20orf99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 3. | |||||
|
DUS3L_HUMAN
|
||||||
| NC score | 0.081472 (rank : 43) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96G46, Q96HM5, Q9BSU4, Q9H877, Q9NPR1 | Gene names | DUS3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
TRM1_HUMAN
|
||||||
| NC score | 0.077025 (rank : 44) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NXH9, O76103, Q548Y5, Q8WVA6 | Gene names | TRMT1 | |||
|
Domain Architecture |

|
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.072259 (rank : 45) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.066773 (rank : 46) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.064902 (rank : 47) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
DUS3L_MOUSE
|
||||||
| NC score | 0.063947 (rank : 48) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91XI1, Q3TZQ1, Q7TT12 | Gene names | Dus3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-dihydrouridine synthase 3-like (EC 1.-.-.-). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.057157 (rank : 49) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
TRM1_MOUSE
|
||||||
| NC score | 0.052243 (rank : 50) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TX08, Q3TBY4 | Gene names | Trmt1, D8Ertd812e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N(2),N(2)-dimethylguanosine tRNA methyltransferase (EC 2.1.1.32) (tRNA(guanine-26,N(2)-N(2)) methyltransferase) (tRNA 2,2- dimethylguanosine-26 methyltransferase) (tRNA(m(2,2)G26)dimethyltransferase). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.052182 (rank : 51) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
PARPT_MOUSE
|
||||||
| NC score | 0.052068 (rank : 52) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C1B2, Q3UD50, Q8C032 | Gene names | Tiparp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.051947 (rank : 53) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.050151 (rank : 54) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.049183 (rank : 55) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.049136 (rank : 56) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.045233 (rank : 57) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.036209 (rank : 58) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.034735 (rank : 59) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZCH12_HUMAN
|
||||||
| NC score | 0.034670 (rank : 60) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PEW1, Q6PID5, Q8N1C1 | Gene names | ZCCHC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 12. | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.022200 (rank : 61) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.021737 (rank : 62) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.021299 (rank : 63) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
ERO1B_HUMAN
|
||||||
| NC score | 0.016484 (rank : 64) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YB8, Q8IZ11, Q9NR62 | Gene names | ERO1LB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
ERO1B_MOUSE
|
||||||
| NC score | 0.016440 (rank : 65) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.015207 (rank : 66) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
ECT2_HUMAN
|
||||||
| NC score | 0.013818 (rank : 67) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8V3, Q9NSV8, Q9NVW9 | Gene names | ECT2 | |||
|
Domain Architecture |

|
|||||
| Description | ECT2 protein (Epithelial cell-transforming sequence 2 oncogene). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.008156 (rank : 68) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.001832 (rank : 69) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
ZN615_HUMAN
|
||||||
| NC score | -0.003211 (rank : 70) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N8J6 | Gene names | ZNF615 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 615. | |||||