Please be patient as the page loads
|
ERO1B_MOUSE
|
||||||
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ERO1B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999511 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YB8, Q8IZ11, Q9NR62 | Gene names | ERO1LB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
ERO1B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
ERO1A_HUMAN
|
||||||
| θ value | 1.23469e-174 (rank : 3) | NC score | 0.992177 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HE7, Q7LD45, Q9P1Q9, Q9UKV6 | Gene names | ERO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein alpha precursor (EC 1.8.4.-) (ERO1-Lalpha) (Oxidoreductin-1-Lalpha) (Endoplasmic oxidoreductin-1-like protein) (ERO1-L). | |||||
|
ERO1A_MOUSE
|
||||||
| θ value | 7.01099e-170 (rank : 4) | NC score | 0.991927 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R180, Q9CV47, Q9QY03 | Gene names | Ero1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein alpha precursor (EC 1.8.4.-) (ERO1-Lalpha) (Oxidoreductin-1-Lalpha) (Endoplasmic oxidoreductin-1-like protein) (ERO1-L). | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 5) | NC score | 0.033860 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
LRRK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 6) | NC score | 0.004368 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 1359 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5S006, Q8BWG7, Q8BZJ6, Q8CI84, Q8K062 | Gene names | Lrrk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
XAB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.036028 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCS7, Q8TET6, Q96HB0, Q96IW0, Q9NRG6, Q9ULP3 | Gene names | XAB2, HCNP, KIAA1177 | |||
|
Domain Architecture |
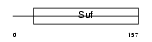
|
|||||
| Description | XPA-binding protein 2 (HCNP protein). | |||||
|
XAB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.036200 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCD2, Q8VDT5, Q9CVD8 | Gene names | Xab2 | |||
|
Domain Architecture |
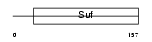
|
|||||
| Description | XPA-binding protein 2. | |||||
|
CPSF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.016440 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
LRRK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.003130 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||
|
ERO1B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
ERO1B_HUMAN
|
||||||
| NC score | 0.999511 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YB8, Q8IZ11, Q9NR62 | Gene names | ERO1LB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
ERO1A_HUMAN
|
||||||
| NC score | 0.992177 (rank : 3) | θ value | 1.23469e-174 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96HE7, Q7LD45, Q9P1Q9, Q9UKV6 | Gene names | ERO1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein alpha precursor (EC 1.8.4.-) (ERO1-Lalpha) (Oxidoreductin-1-Lalpha) (Endoplasmic oxidoreductin-1-like protein) (ERO1-L). | |||||
|
ERO1A_MOUSE
|
||||||
| NC score | 0.991927 (rank : 4) | θ value | 7.01099e-170 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R180, Q9CV47, Q9QY03 | Gene names | Ero1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein alpha precursor (EC 1.8.4.-) (ERO1-Lalpha) (Oxidoreductin-1-Lalpha) (Endoplasmic oxidoreductin-1-like protein) (ERO1-L). | |||||
|
XAB2_MOUSE
|
||||||
| NC score | 0.036200 (rank : 5) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCD2, Q8VDT5, Q9CVD8 | Gene names | Xab2 | |||
|
Domain Architecture |

|
|||||
| Description | XPA-binding protein 2. | |||||
|
XAB2_HUMAN
|
||||||
| NC score | 0.036028 (rank : 6) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCS7, Q8TET6, Q96HB0, Q96IW0, Q9NRG6, Q9ULP3 | Gene names | XAB2, HCNP, KIAA1177 | |||
|
Domain Architecture |

|
|||||
| Description | XPA-binding protein 2 (HCNP protein). | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.033860 (rank : 7) | θ value | 1.38821 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
CPSF4_HUMAN
|
||||||
| NC score | 0.016440 (rank : 8) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95639, Q86TF8, Q9BTW6 | Gene names | CPSF4, CPSF30, NAR, NEB1 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (NS1 effector domain-binding protein 1) (Neb-1) (No arches homolog). | |||||
|
LRRK2_MOUSE
|
||||||
| NC score | 0.004368 (rank : 9) | θ value | 3.0926 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 1359 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5S006, Q8BWG7, Q8BZJ6, Q8CI84, Q8K062 | Gene names | Lrrk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
LRRK2_HUMAN
|
||||||
| NC score | 0.003130 (rank : 10) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||