Please be patient as the page loads
|
PCX3_MOUSE
|
||||||
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PCX1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.922885 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
CN135_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 3) | NC score | 0.440146 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63HM2, Q9BQG8, Q9H9F2 | Gene names | C14orf135, FBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf135 precursor (Hepatitis C virus F protein-binding protein 2) (HCV F protein-binding protein 2). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 4) | NC score | 0.088566 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 5) | NC score | 0.094390 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.060036 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.017555 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.059255 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.062091 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.041972 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.050369 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.058431 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.068141 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.028415 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
IKKB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.014736 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14920, O75327 | Gene names | IKBKB, IKKB | |||
|
Domain Architecture |
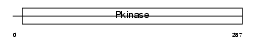
|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.082832 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.044806 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.061065 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.059614 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
OR6V1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.002406 (rank : 181) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N148 | Gene names | OR6V1 | |||
|
Domain Architecture |
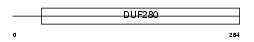
|
|||||
| Description | Olfactory receptor 6V1. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.062233 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.038807 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
IKKB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.013923 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88351, Q9R1J6 | Gene names | Ikbkb, Ikkb | |||
|
Domain Architecture |
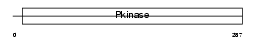
|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.046279 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
PIGQ_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.052492 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.051679 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CD19_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.046464 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |
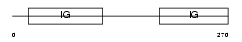
|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.039697 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.059571 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.043067 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.060258 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.068076 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.025835 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.026596 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.060801 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.024210 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.096615 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DEDD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.035999 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.059556 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
E2F4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.033884 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.046361 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
GPDM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.047013 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.030284 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
LZTS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.025636 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.020676 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.082487 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.046282 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
ERO1B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.033860 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
GPSM2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.034255 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.034242 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
LORI_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.055002 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.044971 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
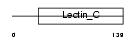
C1QR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.012415 (rank : 167) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.014610 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.023623 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.064066 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.024246 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
RANB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.039139 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
SSDH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.014443 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.030420 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.033205 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ES8L2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.016922 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.038078 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.053268 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.039252 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.014805 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ACHA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.007333 (rank : 177) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15822, Q9HAQ3 | Gene names | CHRNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-2 precursor. | |||||
|
ANS4B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.017407 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ATS15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.007584 (rank : 176) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE58 | Gene names | ADAMTS15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 15) (ADAM-TS 15) (ADAM-TS15). | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.040943 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.039866 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.036385 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
GPC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.017861 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZF2 | Gene names | Gpc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-1 precursor. | |||||
|
GPC5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.019810 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CAL5 | Gene names | Gpc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-5 precursor. | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.039293 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
PROP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.012972 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.021539 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.013783 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.061442 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GTR6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.013435 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGQ3, Q5SXD7 | Gene names | SLC2A6, GLUT9 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 6 (Glucose transporter type 6) (Glucose transporter type 9). | |||||
|
GTSE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.032794 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.008243 (rank : 173) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MK04_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.006003 (rank : 179) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.048396 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.032578 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NOS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.015702 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.017741 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
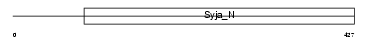
SAC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.024878 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |
|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.038502 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SRPK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.009914 (rank : 170) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
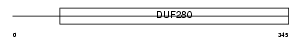
ADA1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.000552 (rank : 182) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35368 | Gene names | ADRA1B | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-1B adrenergic receptor (Alpha 1B-adrenoceptor) (Alpha 1B- adrenoreceptor). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.012770 (rank : 164) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
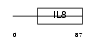
CCL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.012535 (rank : 165) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13236, P22617, Q13704, Q3SXL8, Q6FGI8 | Gene names | CCL4, LAG1, MIP1B, SCYA4 | |||
|
Domain Architecture |
|
|||||
| Description | Small inducible cytokine A4 precursor (CCL4) (Macrophage inflammatory protein 1-beta) (MIP-1-beta) (MIP-1-beta(1-69)) (T-cell activation protein 2) (ACT-2) (PAT 744) (H400) (SIS-gamma) (Lymphocyte activation gene 1 protein) (LAG-1) (HC21) (G-26 T-lymphocyte-secreted protein) [Contains: MIP-1-beta(3-69)]. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.019355 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
EPN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.030243 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
ERO1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.027084 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YB8, Q8IZ11, Q9NR62 | Gene names | ERO1LB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.021145 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.030230 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.041173 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.023254 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.020835 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.012429 (rank : 166) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.008962 (rank : 171) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ADPN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.017380 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NST1, Q6P1A1, Q96CB4 | Gene names | PNPLA3, ADPN, C22orf20 | |||
|
Domain Architecture |
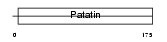
|
|||||
| Description | Adiponutrin (iPLA2-epsilon) (Calcium-independent phospholipase A2- epsilon) (Patatin-like phospholipase domain-containing protein 3) [Includes: Triacylglycerol lipase (EC 3.1.1.3); Acylglycerol O- acyltransferase (EC 2.3.1.-)]. | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.022373 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.045269 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.049656 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
CT128_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.019830 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
CT160_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.017358 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUG4, Q5JYR9, Q8N5F1, Q8N6G8, Q96MD5 | Gene names | C20orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf160. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.004460 (rank : 180) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.049612 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.044234 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.018063 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TERT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.017976 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14746, O14783, Q2XS35, Q8N6C3 | Gene names | TERT, EST2, TCS1, TRT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomerase reverse transcriptase (EC 2.7.7.49) (Telomerase catalytic subunit) (HEST2) (Telomerase-associated protein 2) (TP2). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.008948 (rank : 172) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
ZN746_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | -0.000214 (rank : 183) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NUN9 | Gene names | ZNF746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||
|
APR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.027154 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5M1, Q6NTD6 | Gene names | APR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-related protein 2 (APR-2). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.031973 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
COJA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.030461 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.010014 (rank : 169) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.015875 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
FOXJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.011528 (rank : 168) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |
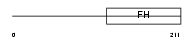
|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
FREA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.015471 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.006083 (rank : 178) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
IDUA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.020559 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48441 | Gene names | Idua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-L-iduronidase precursor (EC 3.2.1.76). | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.020208 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
MYL6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.007634 (rank : 174) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |
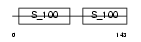
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.007634 (rank : 175) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |
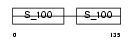
|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.013794 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.041598 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.023904 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SAC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.020568 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.049746 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TAU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.042708 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050955 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.056166 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.061266 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.063242 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.059122 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.076001 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.078410 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.050776 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.069036 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.069491 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051581 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.053722 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.057472 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.052867 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.065232 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.064715 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072811 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.060523 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.095392 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.093519 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.050227 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.101089 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.113249 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.078401 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.072428 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.052500 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.077297 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.073552 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.056313 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.055919 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052683 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.052578 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.052735 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052488 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.072342 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.053025 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.056462 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.054939 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.052228 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.073888 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.078515 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.076901 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.051845 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.058969 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.068910 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.065220 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.064227 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060679 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.053493 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.922885 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
CN135_HUMAN
|
||||||
| NC score | 0.440146 (rank : 3) | θ value | 3.89403e-11 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63HM2, Q9BQG8, Q9H9F2 | Gene names | C14orf135, FBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf135 precursor (Hepatitis C virus F protein-binding protein 2) (HCV F protein-binding protein 2). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.113249 (rank : 4) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.101089 (rank : 5) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.096615 (rank : 6) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.095392 (rank : 7) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.094390 (rank : 8) | θ value | 5.44631e-05 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.093519 (rank : 9) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.088566 (rank : 10) | θ value | 6.43352e-06 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.082832 (rank : 11) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.082487 (rank : 12) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.078515 (rank : 13) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.078410 (rank : 14) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.078401 (rank : 15) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.077297 (rank : 16) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.076901 (rank : 17) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.076001 (rank : 18) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.073888 (rank : 19) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.073552 (rank : 20) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.072811 (rank : 21) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.072428 (rank : 22) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.072342 (rank : 23) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.069491 (rank : 24) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.069036 (rank : 25) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.068910 (rank : 26) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.068141 (rank : 27) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.068076 (rank : 28) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.065232 (rank : 29) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.065220 (rank : 30) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.064715 (rank : 31) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.064227 (rank : 32) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.064066 (rank : 33) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.063242 (rank : 34) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.062233 (rank : 35) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.062091 (rank : 36) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.061442 (rank : 37) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.061266 (rank : 38) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.061065 (rank : 39) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.060801 (rank : 40) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.060679 (rank : 41) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.060523 (rank : 42) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.060258 (rank : 43) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.060036 (rank : 44) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.059614 (rank : 45) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.059571 (rank : 46) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.059556 (rank : 47) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.059255 (rank : 48) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.059122 (rank : 49) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.058969 (rank : 50) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.058431 (rank : 51) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.057472 (rank : 52) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.056462 (rank : 53) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.056313 (rank : 54) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.056166 (rank : 55) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.055919 (rank : 56) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.055002 (rank : 57) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.054939 (rank : 58) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.053722 (rank : 59) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.053493 (rank : 60) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.053268 (rank : 61) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.053025 (rank : 62) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.052867 (rank : 63) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.052735 (rank : 64) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.052683 (rank : 65) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.052578 (rank : 66) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.052500 (rank : 67) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PIGQ_HUMAN
|
||||||
| NC score | 0.052492 (rank : 68) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRB3, O14927, Q96G00, Q96S22, Q9UJH4 | Gene names | PIGQ, GPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q (EC 2.4.1.198) (Phosphatidylinositol-glycan biosynthesis class Q protein) (PIG-Q) (N-acetylglucosamyl transferase component GPI1). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.052488 (rank : 69) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.052228 (rank : 70) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.051845 (rank : 71) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.051679 (rank : 72) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.051581 (rank : 73) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.050955 (rank : 74) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.050776 (rank : 75) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.050369 (rank : 76) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.050227 (rank : 77) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.049746 (rank : 78) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.049656 (rank : 79) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.049612 (rank : 80) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.048396 (rank : 81) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.047013 (rank : 82) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
CD19_HUMAN
|
||||||
| NC score | 0.046464 (rank : 83) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |

|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.046361 (rank : 84) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.046282 (rank : 85) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.046279 (rank : 86) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.045269 (rank : 87) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.044971 (rank : 88) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.044806 (rank : 89) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.044234 (rank : 90) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.043067 (rank : 91) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.042708 (rank : 92) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.041972 (rank : 93) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.041598 (rank : 94) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.041173 (rank : 95) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.040943 (rank : 96) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.039866 (rank : 97) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.039697 (rank : 98) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.039293 (rank : 99) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.039252 (rank : 100) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RANB3_MOUSE
|
||||||
| NC score | 0.039139 (rank : 101) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CT10, Q3TIS4, Q3U2G4, Q3UYG7 | Gene names | Ranbp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.038807 (rank : 102) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.038502 (rank : 103) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.038078 (rank : 104) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.036385 (rank : 105) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
DEDD2_MOUSE
|
||||||
| NC score | 0.035999 (rank : 106) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
GPSM2_HUMAN
|
||||||
| NC score | 0.034255 (rank : 107) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P81274, Q8N0Z5 | Gene names | GPSM2, LGN | |||
|
Domain Architecture |

|
|||||
| Description | G-protein signaling modulator 2 (Mosaic protein LGN). | |||||
|
GPSM2_MOUSE
|
||||||
| NC score | 0.034242 (rank : 108) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDU0, Q8BLX3 | Gene names | Gpsm2, Lgn, Pins | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein signaling modulator 2 (Pins homolog). | |||||
|
E2F4_HUMAN
|
||||||
| NC score | 0.033884 (rank : 109) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
ERO1B_MOUSE
|
||||||
| NC score | 0.033860 (rank : 110) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R2E9 | Gene names | Ero1lb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.033205 (rank : 111) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GTSE1_HUMAN
|
||||||
| NC score | 0.032794 (rank : 112) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYZ3, Q53GX5, Q5R3I6, Q6DHX4, Q9BRE0, Q9UGZ9, Q9Y557 | Gene names | GTSE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (B99 homolog). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.032578 (rank : 113) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.031973 (rank : 114) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.030461 (rank : 115) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.030420 (rank : 116) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.030284 (rank : 117) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
EPN3_MOUSE
|
||||||
| NC score | 0.030243 (rank : 118) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91W69, Q9CV55 | Gene names | Epn3 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-3 (EPS-15-interacting protein 3). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.030230 (rank : 119) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.028415 (rank : 120) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
APR2_HUMAN
|
||||||
| NC score | 0.027154 (rank : 121) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5M1, Q6NTD6 | Gene names | APR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-related protein 2 (APR-2). | |||||
|
ERO1B_HUMAN
|
||||||
| NC score | 0.027084 (rank : 122) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86YB8, Q8IZ11, Q9NR62 | Gene names | ERO1LB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERO1-like protein beta precursor (EC 1.8.4.-) (ERO1-Lbeta) (Oxidoreductin-1-Lbeta) (Endoplasmic oxidoreductin-1-like protein B). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.026596 (rank : 123) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.025835 (rank : 124) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
LZTS2_HUMAN
|
||||||
| NC score | 0.025636 (rank : 125) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
SAC3_HUMAN
|
||||||
| NC score | 0.024878 (rank : 126) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92562, Q53H49, Q5TCS6 | Gene names | SAC3, KIAA0274 | |||
|
Domain Architecture |

|
|||||
| Description | SAC domain-containing protein 3. | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.024246 (rank : 127) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.024210 (rank : 128) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.023904 (rank : 129) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.023623 (rank : 130) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.023254 (rank : 131) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.022373 (rank : 132) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.021539 (rank : 133) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.021145 (rank : 134) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.020835 (rank : 135) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.020676 (rank : 136) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SAC3_MOUSE
|
||||||
| NC score | 0.020568 (rank : 137) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
IDUA_MOUSE
|
||||||
| NC score | 0.020559 (rank : 138) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48441 | Gene names | Idua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-L-iduronidase precursor (EC 3.2.1.76). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.020208 (rank : 139) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
CT128_HUMAN
|
||||||
| NC score | 0.019830 (rank : 140) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
GPC5_MOUSE
|
||||||
| NC score | 0.019810 (rank : 141) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CAL5 | Gene names | Gpc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-5 precursor. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.019355 (rank : 142) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.018063 (rank : 143) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TERT_HUMAN
|
||||||
| NC score | 0.017976 (rank : 144) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14746, O14783, Q2XS35, Q8N6C3 | Gene names | TERT, EST2, TCS1, TRT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomerase reverse transcriptase (EC 2.7.7.49) (Telomerase catalytic subunit) (HEST2) (Telomerase-associated protein 2) (TP2). | |||||
|
GPC1_MOUSE
|
||||||
| NC score | 0.017861 (rank : 145) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZF2 | Gene names | Gpc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glypican-1 precursor. | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.017741 (rank : 146) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.017555 (rank : 147) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ANS4B_HUMAN
|
||||||
| NC score | 0.017407 (rank : 148) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8V4 | Gene names | ANKS4B, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 4B (Harmonin- interacting ankyrin repeat-containing protein) (Harp). | |||||
|
ADPN_HUMAN
|
||||||
| NC score | 0.017380 (rank : 149) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NST1, Q6P1A1, Q96CB4 | Gene names | PNPLA3, ADPN, C22orf20 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponutrin (iPLA2-epsilon) (Calcium-independent phospholipase A2- epsilon) (Patatin-like phospholipase domain-containing protein 3) [Includes: Triacylglycerol lipase (EC 3.1.1.3); Acylglycerol O- acyltransferase (EC 2.3.1.-)]. | |||||
|
CT160_HUMAN
|
||||||
| NC score | 0.017358 (rank : 150) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUG4, Q5JYR9, Q8N5F1, Q8N6G8, Q96MD5 | Gene names | C20orf160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf160. | |||||
|
ES8L2_HUMAN
|
||||||
| NC score | 0.016922 (rank : 151) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.015875 (rank : 152) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
NOS1_HUMAN
|
||||||
| NC score | 0.015702 (rank : 153) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29475 | Gene names | NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, brain (EC 1.14.13.39) (NOS type I) (Neuronal NOS) (N-NOS) (nNOS) (Constitutive NOS) (NC-NOS) (bNOS). | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.015471 (rank : 154) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.014805 (rank : 155) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
IKKB_HUMAN
|
||||||
| NC score | 0.014736 (rank : 156) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14920, O75327 | Gene names | IKBKB, IKKB | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.014610 (rank : 157) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
SSDH_MOUSE
|
||||||
| NC score | 0.014443 (rank : 158) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWF0 | Gene names | Aldh5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate semialdehyde dehydrogenase, mitochondrial precursor (EC 1.2.1.24) (NAD(+)-dependent succinic semialdehyde dehydrogenase). | |||||
|
IKKB_MOUSE
|
||||||
| NC score | 0.013923 (rank : 159) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88351, Q9R1J6 | Gene names | Ikbkb, Ikkb | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa B kinase subunit beta (EC 2.7.11.10) (I-kappa-B-kinase beta) (IkBKB) (IKK-beta) (IKK-B) (I-kappa-B kinase 2) (IKK2) (Nuclear factor NF-kappa-B inhibitor kinase beta) (NFKBIKB). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.013794 (rank : 160) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.013783 (rank : 161) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
GTR6_HUMAN
|
||||||
| NC score | 0.013435 (rank : 162) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGQ3, Q5SXD7 | Gene names | SLC2A6, GLUT9 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 2, facilitated glucose transporter member 6 (Glucose transporter type 6) (Glucose transporter type 9). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.012972 (rank : 163) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.012770 (rank : 164) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CCL4_HUMAN
|
||||||
| NC score | 0.012535 (rank : 165) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13236, P22617, Q13704, Q3SXL8, Q6FGI8 | Gene names | CCL4, LAG1, MIP1B, SCYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Small inducible cytokine A4 precursor (CCL4) (Macrophage inflammatory protein 1-beta) (MIP-1-beta) (MIP-1-beta(1-69)) (T-cell activation protein 2) (ACT-2) (PAT 744) (H400) (SIS-gamma) (Lymphocyte activation gene 1 protein) (LAG-1) (HC21) (G-26 T-lymphocyte-secreted protein) [Contains: MIP-1-beta(3-69)]. | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.012429 (rank : 166) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.012415 (rank : 167) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
FOXJ1_MOUSE
|
||||||
| NC score | 0.011528 (rank : 168) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.010014 (rank : 169) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
SRPK1_MOUSE
|
||||||
| NC score | 0.009914 (rank : 170) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.008962 (rank : 171) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.008948 (rank : 172) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.008243 (rank : 173) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MYL6_HUMAN
|
||||||
| NC score | 0.007634 (rank : 174) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60660, P16475, P24572, P24573, Q12790, Q6IAZ0, Q6IPY5 | Gene names | MYL6 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
MYL6_MOUSE
|
||||||
| NC score | 0.007634 (rank : 175) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60605 | Gene names | Myl6, Myln | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 6 (Smooth muscle and nonmuscle myosin light chain alkali 6) (Myosin light chain alkali 3) (Myosin light chain 3) (MLC-3) (LC17). | |||||
|
ATS15_HUMAN
|
||||||
| NC score | 0.007584 (rank : 176) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE58 | Gene names | ADAMTS15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 15) (ADAM-TS 15) (ADAM-TS15). | |||||
|
ACHA2_HUMAN
|
||||||
| NC score | 0.007333 (rank : 177) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15822, Q9HAQ3 | Gene names | CHRNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal acetylcholine receptor protein subunit alpha-2 precursor. | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.006083 (rank : 178) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
MK04_MOUSE
|
||||||
| NC score | 0.006003 (rank : 179) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.004460 (rank : 180) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
OR6V1_HUMAN
|
||||||
| NC score | 0.002406 (rank : 181) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N148 | Gene names | OR6V1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 6V1. | |||||
|
ADA1B_HUMAN
|
||||||
| NC score | 0.000552 (rank : 182) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35368 | Gene names | ADRA1B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1B adrenergic receptor (Alpha 1B-adrenoceptor) (Alpha 1B- adrenoreceptor). | |||||
|
ZN746_HUMAN
|
||||||
| NC score | -0.000214 (rank : 183) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NUN9 | Gene names | ZNF746 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 746. | |||||