Please be patient as the page loads
|
E2F4_HUMAN
|
||||||
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
E2F4_HUMAN
|
||||||
| θ value | 1.71888e-184 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 188 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
E2F5_HUMAN
|
||||||
| θ value | 6.73165e-80 (rank : 2) | NC score | 0.942360 (rank : 2) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15329, Q16601, Q92756 | Gene names | E2F5 | |||
|
Domain Architecture |
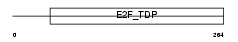
|
|||||
| Description | Transcription factor E2F5 (E2F-5). | |||||
|
E2F5_MOUSE
|
||||||
| θ value | 4.08394e-77 (rank : 3) | NC score | 0.942132 (rank : 3) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61502 | Gene names | E2f5 | |||
|
Domain Architecture |
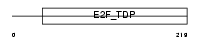
|
|||||
| Description | Transcription factor E2F5 (E2F-5). | |||||
|
E2F2_MOUSE
|
||||||
| θ value | 2.58786e-39 (rank : 4) | NC score | 0.866889 (rank : 4) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56931, Q8BID0 | Gene names | E2f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
E2F3_MOUSE
|
||||||
| θ value | 5.96599e-36 (rank : 5) | NC score | 0.862374 (rank : 6) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F3_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 6) | NC score | 0.862133 (rank : 8) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |
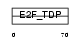
|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F2_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 7) | NC score | 0.862009 (rank : 9) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
E2F1_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 8) | NC score | 0.846948 (rank : 10) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01094, Q13143, Q92768 | Gene names | E2F1, RBBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1) (Retinoblastoma-binding protein 3) (RBBP-3) (PRB-binding protein E2F-1) (PBR3) (Retinoblastoma-associated protein 1) (RBAP-1). | |||||
|
E2F6_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 9) | NC score | 0.862174 (rank : 7) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54917 | Gene names | E2f6 | |||
|
Domain Architecture |
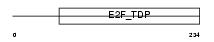
|
|||||
| Description | Transcription factor E2F6 (E2F-6) (E2F-binding site-modulating activity protein) (EMA). | |||||
|
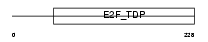
E2F6_HUMAN
|
||||||
| θ value | 1.02001e-27 (rank : 10) | NC score | 0.862689 (rank : 5) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75461, O60544, Q7Z2H6 | Gene names | E2F6 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor E2F6 (E2F-6). | |||||
|
E2F1_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 11) | NC score | 0.837497 (rank : 11) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61501, Q80VZ3 | Gene names | E2f1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 12) | NC score | 0.077546 (rank : 18) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 13) | NC score | 0.086564 (rank : 15) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.084343 (rank : 16) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.041645 (rank : 74) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.066247 (rank : 25) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.066349 (rank : 24) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.021779 (rank : 142) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.066756 (rank : 22) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.066462 (rank : 23) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.038107 (rank : 85) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.056614 (rank : 47) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.065161 (rank : 28) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.036816 (rank : 91) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.056533 (rank : 48) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.008228 (rank : 174) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.058497 (rank : 39) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
URFB1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.057297 (rank : 44) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.065872 (rank : 26) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.052945 (rank : 54) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.048704 (rank : 60) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.058525 (rank : 38) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.045655 (rank : 66) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.077891 (rank : 17) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.026716 (rank : 123) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
GBX1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.007956 (rank : 177) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14549 | Gene names | GBX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein GBX-1 (Gastrulation and brain-specific homeobox protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.035437 (rank : 95) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.067410 (rank : 21) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.051742 (rank : 56) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SHB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.022850 (rank : 134) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15464, Q504U5, Q5VUM8 | Gene names | SHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein B. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.061617 (rank : 34) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.054594 (rank : 52) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.017213 (rank : 149) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.038635 (rank : 83) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.037023 (rank : 89) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.027586 (rank : 121) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.036239 (rank : 92) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.073458 (rank : 19) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.065719 (rank : 27) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.055365 (rank : 51) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.043573 (rank : 71) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
FSBP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.036077 (rank : 93) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKE5, Q8BJX8 | Gene names | Fsbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen silencer-binding protein. | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.016646 (rank : 150) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.046177 (rank : 65) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.065035 (rank : 29) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.051560 (rank : 57) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.010318 (rank : 168) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.060533 (rank : 36) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.014500 (rank : 157) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.031524 (rank : 105) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
AKTS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.038759 (rank : 81) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D1F4 | Gene names | Akt1s1, Pras | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (Proline-rich AKT substrate). | |||||
|
AP180_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.044406 (rank : 67) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
I12R2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.018627 (rank : 146) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99665 | Gene names | IL12RB2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-2 chain precursor (IL-12 receptor beta-2) (IL-12R-beta2). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.051162 (rank : 59) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.062823 (rank : 32) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.033884 (rank : 99) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.058379 (rank : 40) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.028009 (rank : 120) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.027090 (rank : 122) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.047238 (rank : 64) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TAF10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.040012 (rank : 79) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12962, O00703, Q13175 | Gene names | TAF10, TAF2H, TAFII30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (TAFII30) (STAF28). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.015826 (rank : 154) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 1.06291 (rank : 73) | NC score | 0.002532 (rank : 190) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.052930 (rank : 55) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
K2002_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.021901 (rank : 141) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H792, Q6ZS78, Q8NCM3 | Gene names | KIAA2002 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized tyrosine-protein kinase KIAA2002 (EC 2.7.10.2). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.061634 (rank : 33) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.043943 (rank : 68) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.039681 (rank : 80) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.033433 (rank : 101) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
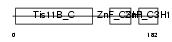
TISD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.025641 (rank : 126) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |
|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
YTHD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.022511 (rank : 137) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
CO1A2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.031899 (rank : 104) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.021570 (rank : 143) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
EP400_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.025677 (rank : 125) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.028947 (rank : 112) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
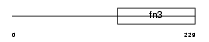
IL2RB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.025568 (rank : 127) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16297, Q3TZT2 | Gene names | Il2rb | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.035672 (rank : 94) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.011131 (rank : 167) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.037142 (rank : 88) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.015769 (rank : 155) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.013039 (rank : 160) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.025701 (rank : 124) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.043914 (rank : 69) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
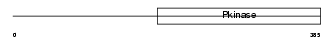
IRAK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.005691 (rank : 182) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |
|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.006389 (rank : 181) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.057825 (rank : 42) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.028396 (rank : 116) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.048413 (rank : 62) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.018215 (rank : 148) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.051194 (rank : 58) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.055646 (rank : 50) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.021025 (rank : 144) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
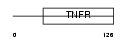
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
ADA12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.004766 (rank : 183) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.035198 (rank : 97) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.022613 (rank : 135) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CR025_HUMAN
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.024929 (rank : 129) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.034271 (rank : 98) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.058133 (rank : 41) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.029663 (rank : 110) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NF2L1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.028193 (rank : 119) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
NF2L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.028280 (rank : 117) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.020264 (rank : 145) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.028262 (rank : 118) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.033630 (rank : 100) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.047347 (rank : 63) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.025092 (rank : 128) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.048454 (rank : 61) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.040819 (rank : 76) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.040114 (rank : 78) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
ST2B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.008139 (rank : 175) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00204, O00205, O75814 | Gene names | SULT2B1, HSST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase family cytosolic 2B member 1 (EC 2.8.2.2) (Sulfotransferase 2B1) (Alcohol sulfotransferase) (Hydroxysteroid sulfotransferase 2). | |||||
|
SYNP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.030468 (rank : 108) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
TAF6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.030990 (rank : 107) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49848 | Gene names | TAF6, TAF2E, TAFII70 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 6 (Transcription initiation factor TFIID 70 kDa subunit) (TAF(II)70) (TAFII-70) (TAFII- 80) (TAFII80). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.032179 (rank : 103) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TIE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | -0.000265 (rank : 193) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.016481 (rank : 151) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.030190 (rank : 109) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.029139 (rank : 111) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.043664 (rank : 70) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.022301 (rank : 139) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.036872 (rank : 90) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.032711 (rank : 102) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.022604 (rank : 136) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.038488 (rank : 84) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.008749 (rank : 171) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.008565 (rank : 172) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MUC15_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.022472 (rank : 138) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C6Z1, Q3UQC4, Q6XYQ9, Q7TMH6, Q8CE82 | Gene names | Muc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
PGLB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.003730 (rank : 187) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99645, Q8NEJ5 | Gene names | DSPG3, PGLB | |||
|
Domain Architecture |

|
|||||
| Description | Dermatan sulfate proteoglycan 3 precursor (Epiphycan) (Small chondroitin/dermatan sulfate proteoglycan) (Proteoglycan-Lb) (PG-Lb). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.023786 (rank : 131) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
SCRT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | -0.000749 (rank : 196) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99M85 | Gene names | Scrt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor scratch 1 (Scratch homolog 1 zinc finger protein) (Scratch 1) (SCRT) (mScrt). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.041421 (rank : 75) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.006941 (rank : 180) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.022283 (rank : 140) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.016315 (rank : 152) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.038701 (rank : 82) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.031361 (rank : 106) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.028861 (rank : 113) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
DBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.011333 (rank : 165) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
EGR4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | -0.000146 (rank : 192) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.043432 (rank : 72) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
FOXF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.004050 (rank : 185) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |
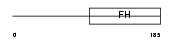
|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
HRSL5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.011435 (rank : 164) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96KN8 | Gene names | HRASLS5, HRLP5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
KLF3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | -0.000505 (rank : 195) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60980 | Gene names | Klf3, Bklf | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 3 (Basic krueppel-like factor) (CACCC-box-binding protein BKLF) (TEF-2). | |||||
|
LAMB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.012325 (rank : 161) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.037196 (rank : 87) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.004396 (rank : 184) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.001061 (rank : 191) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.071170 (rank : 20) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.057486 (rank : 43) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PHAR3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.013990 (rank : 158) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96KR7, Q707P6, Q9H4T4 | Gene names | PHACTR3, C20orf101, SCAPIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
PTPRJ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.002580 (rank : 189) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
SBK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.002865 (rank : 188) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8QZX0 | Gene names | Sbk1, Sbk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SBK1 (EC 2.7.11.1) (SH3-binding kinase 1). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.023614 (rank : 132) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | 0.023576 (rank : 133) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.016271 (rank : 153) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SOX3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.007135 (rank : 179) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 166) | NC score | 0.042547 (rank : 73) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
TDP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 167) | NC score | 0.136925 (rank : 13) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14186, Q8IZL5 | Gene names | TFDP1, DP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-1 (E2F dimerization partner 1) (DRTF1- polypeptide 1) (DRTF1). | |||||
|
TDP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 168) | NC score | 0.139445 (rank : 12) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08639 | Gene names | Tfdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-1 (E2F dimerization partner 1) (DRTF1- polypeptide 1). | |||||
|
TDP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 169) | NC score | 0.108149 (rank : 14) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14188, Q13331, Q14187, Q6R754, Q8WU88 | Gene names | TFDP2, DP2 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor Dp-2 (E2F dimerization partner 2). | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.028511 (rank : 115) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
CENPU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.018437 (rank : 147) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
CO039_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.040381 (rank : 77) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
CRTC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.028616 (rank : 114) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
CS007_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.037974 (rank : 86) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
FBXL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.008956 (rank : 170) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C2S5, Q3TWC9, Q80XI5, Q8BGF5, Q8BNL3, Q8C3Q8 | Gene names | Fbxl5 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 5 (F-box and leucine-rich repeat protein 5). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.012264 (rank : 162) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
FOXGB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.003921 (rank : 186) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.013101 (rank : 159) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
LSP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.015274 (rank : 156) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.008518 (rank : 173) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.007996 (rank : 176) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | -0.000309 (rank : 194) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.007955 (rank : 178) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.035406 (rank : 96) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 185) | NC score | 0.010281 (rank : 169) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 186) | NC score | 0.011149 (rank : 166) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 187) | NC score | 0.011930 (rank : 163) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 188) | NC score | 0.024636 (rank : 130) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
CK024_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.057115 (rank : 45) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.061528 (rank : 35) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.064391 (rank : 30) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.063864 (rank : 31) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.056666 (rank : 46) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MYO15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.053808 (rank : 53) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.056000 (rank : 49) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.059298 (rank : 37) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
E2F4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.71888e-184 (rank : 1) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 188 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
E2F5_HUMAN
|
||||||
| NC score | 0.942360 (rank : 2) | θ value | 6.73165e-80 (rank : 2) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15329, Q16601, Q92756 | Gene names | E2F5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F5 (E2F-5). | |||||
|
E2F5_MOUSE
|
||||||
| NC score | 0.942132 (rank : 3) | θ value | 4.08394e-77 (rank : 3) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61502 | Gene names | E2f5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F5 (E2F-5). | |||||
|
E2F2_MOUSE
|
||||||
| NC score | 0.866889 (rank : 4) | θ value | 2.58786e-39 (rank : 4) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56931, Q8BID0 | Gene names | E2f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
E2F6_HUMAN
|
||||||
| NC score | 0.862689 (rank : 5) | θ value | 1.02001e-27 (rank : 10) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75461, O60544, Q7Z2H6 | Gene names | E2F6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F6 (E2F-6). | |||||
|
E2F3_MOUSE
|
||||||
| NC score | 0.862374 (rank : 6) | θ value | 5.96599e-36 (rank : 5) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F6_MOUSE
|
||||||
| NC score | 0.862174 (rank : 7) | θ value | 3.17079e-29 (rank : 9) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54917 | Gene names | E2f6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F6 (E2F-6) (E2F-binding site-modulating activity protein) (EMA). | |||||
|
E2F3_HUMAN
|
||||||
| NC score | 0.862133 (rank : 8) | θ value | 1.01765e-35 (rank : 6) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F2_HUMAN
|
||||||
| NC score | 0.862009 (rank : 9) | θ value | 2.77131e-33 (rank : 7) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
E2F1_HUMAN
|
||||||
| NC score | 0.846948 (rank : 10) | θ value | 1.08979e-29 (rank : 8) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01094, Q13143, Q92768 | Gene names | E2F1, RBBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1) (Retinoblastoma-binding protein 3) (RBBP-3) (PRB-binding protein E2F-1) (PBR3) (Retinoblastoma-associated protein 1) (RBAP-1). | |||||
|
E2F1_MOUSE
|
||||||
| NC score | 0.837497 (rank : 11) | θ value | 2.20094e-22 (rank : 11) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61501, Q80VZ3 | Gene names | E2f1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F1 (E2F-1). | |||||
|
TDP1_MOUSE
|
||||||
| NC score | 0.139445 (rank : 12) | θ value | 6.88961 (rank : 168) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08639 | Gene names | Tfdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-1 (E2F dimerization partner 1) (DRTF1- polypeptide 1). | |||||
|
TDP1_HUMAN
|
||||||
| NC score | 0.136925 (rank : 13) | θ value | 6.88961 (rank : 167) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14186, Q8IZL5 | Gene names | TFDP1, DP1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-1 (E2F dimerization partner 1) (DRTF1- polypeptide 1) (DRTF1). | |||||
|
TDP2_HUMAN
|
||||||
| NC score | 0.108149 (rank : 14) | θ value | 6.88961 (rank : 169) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14188, Q13331, Q14187, Q6R754, Q8WU88 | Gene names | TFDP2, DP2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Dp-2 (E2F dimerization partner 2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.086564 (rank : 15) | θ value | 0.0148317 (rank : 13) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.084343 (rank : 16) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.077891 (rank : 17) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.077546 (rank : 18) | θ value | 0.00134147 (rank : 12) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.073458 (rank : 19) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.071170 (rank : 20) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.067410 (rank : 21) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.066756 (rank : 22) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.066462 (rank : 23) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.066349 (rank : 24) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.066247 (rank : 25) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.065872 (rank : 26) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.065719 (rank : 27) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.065161 (rank : 28) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.065035 (rank : 29) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.064391 (rank : 30) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.063864 (rank : 31) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.062823 (rank : 32) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.061634 (rank : 33) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.061617 (rank : 34) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.061528 (rank : 35) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.060533 (rank : 36) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.059298 (rank : 37) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.058525 (rank : 38) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.058497 (rank : 39) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.058379 (rank : 40) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.058133 (rank : 41) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.057825 (rank : 42) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.057486 (rank : 43) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
URFB1_HUMAN
|
||||||
| NC score | 0.057297 (rank : 44) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.057115 (rank : 45) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.056666 (rank : 46) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.056614 (rank : 47) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.056533 (rank : 48) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.056000 (rank : 49) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.055646 (rank : 50) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.055365 (rank : 51) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.054594 (rank : 52) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MYO15_MOUSE
|
||||||
| NC score | 0.053808 (rank : 53) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QZZ4, O70395, Q9QWL6 | Gene names | Myo15a, Myo15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.052945 (rank : 54) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.052930 (rank : 55) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.051742 (rank : 56) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.051560 (rank : 57) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.051194 (rank : 58) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.051162 (rank : 59) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.048704 (rank : 60) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.048454 (rank : 61) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.048413 (rank : 62) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.047347 (rank : 63) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.047238 (rank : 64) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.046177 (rank : 65) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.045655 (rank : 66) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
AP180_HUMAN
|
||||||
| NC score | 0.044406 (rank : 67) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60641, Q5JX13, Q68DL9, Q6P9D3, Q9NTY7 | Gene names | SNAP91, KIAA0656 | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin coat assembly protein AP180 (Clathrin coat-associated protein AP180) (91 kDa synaptosomal-associated protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.043943 (rank : 68) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.043914 (rank : 69) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.043664 (rank : 70) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.043573 (rank : 71) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.043432 (rank : 72) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.042547 (rank : 73) | θ value | 6.88961 (rank : 166) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.041645 (rank : 74) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.041421 (rank : 75) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.040819 (rank : 76) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.040381 (rank : 77) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.040114 (rank : 78) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
TAF10_HUMAN
|
||||||
| NC score | 0.040012 (rank : 79) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12962, O00703, Q13175 | Gene names | TAF10, TAF2H, TAFII30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 10 (Transcription initiation factor TFIID 30 kDa subunit) (TAF(II)30) (TAFII-30) (TAFII30) (STAF28). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.039681 (rank : 80) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
AKTS1_MOUSE
|
||||||
| NC score | 0.038759 (rank : 81) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D1F4 | Gene names | Akt1s1, Pras | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich AKT1 substrate 1 (Proline-rich AKT substrate). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.038701 (rank : 82) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.038635 (rank : 83) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.038488 (rank : 84) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.038107 (rank : 85) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.037974 (rank : 86) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.037196 (rank : 87) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.037142 (rank : 88) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.037023 (rank : 89) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.036872 (rank : 90) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.036816 (rank : 91) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.036239 (rank : 92) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
FSBP_MOUSE
|
||||||
| NC score | 0.036077 (rank : 93) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKE5, Q8BJX8 | Gene names | Fsbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen silencer-binding protein. | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.035672 (rank : 94) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.035437 (rank : 95) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.035406 (rank : 96) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.035198 (rank : 97) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.034271 (rank : 98) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.033884 (rank : 99) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.033630 (rank : 100) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.033433 (rank : 101) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.032711 (rank : 102) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.032179 (rank : 103) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CO1A2_HUMAN
|
||||||
| NC score | 0.031899 (rank : 104) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.031524 (rank : 105) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.031361 (rank : 106) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
TAF6_HUMAN
|
||||||
| NC score | 0.030990 (rank : 107) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49848 | Gene names | TAF6, TAF2E, TAFII70 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 6 (Transcription initiation factor TFIID 70 kDa subunit) (TAF(II)70) (TAFII-70) (TAFII- 80) (TAFII80). | |||||
|
SYNP2_HUMAN
|
||||||
| NC score | 0.030468 (rank : 108) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UMS6, Q9UK89 | Gene names | SYNPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin) (Genethonin 2). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.030190 (rank : 109) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.029663 (rank : 110) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.029139 (rank : 111) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.028947 (rank : 112) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.028861 (rank : 113) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CRTC1_HUMAN
|
||||||
| NC score | 0.028616 (rank : 114) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6UUV9, O75114, Q6Y3A3, Q7LDZ2, Q8IUL3, Q8IZ34, Q8IZL1, Q8N6W3, Q96AI8, Q9H801 | Gene names | CRTC1, KIAA0616, MECT1, TORC1, WAMTP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Transducer of regulated cAMP response element-binding protein 1) (Transducer of CREB protein 1) (Mucoepidermoid carcinoma translocated protein 1). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.028511 (rank : 115) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.028396 (rank : 116) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
NF2L1_MOUSE
|
||||||
| NC score | 0.028280 (rank : 117) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61985, O70234 | Gene names | Nfe2l1, Nrf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.028262 (rank : 118) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
NF2L1_HUMAN
|
||||||
| NC score | 0.028193 (rank : 119) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14494, Q12877, Q96FN6 | Gene names | NFE2L1, HBZ17, NRF1, TCF11 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 1 (NF-E2-related factor 1) (NFE2-related factor 1) (Nuclear factor, erythroid derived 2, like 1) (Transcription factor 11) (Transcription factor HBZ17) (Transcription factor LCR-F1) (Locus control region-factor 1). | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.028009 (rank : 120) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.027586 (rank : 121) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.027090 (rank : 122) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.026716 (rank : 123) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.025701 (rank : 124) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.025677 (rank : 125) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
TISD_MOUSE
|
||||||
| NC score | 0.025641 (rank : 126) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23949 | Gene names | Zfp36l2, Brf2, Tis11d | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 2 (TIS11D protein). | |||||
|
IL2RB_MOUSE
|
||||||
| NC score | 0.025568 (rank : 127) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16297, Q3TZT2 | Gene names | Il2rb | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-2 receptor beta chain precursor (IL-2 receptor) (P70-75) (High affinity IL-2 receptor subunit beta) (CD122 antigen). | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.025092 (rank : 128) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
CR025_HUMAN
|
||||||
| NC score | 0.024929 (rank : 129) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.024636 (rank : 130) | θ value | 8.99809 (rank : 188) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.023786 (rank : 131) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.023614 (rank : 132) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.023576 (rank : 133) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SHB_HUMAN
|
||||||
| NC score | 0.022850 (rank : 134) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15464, Q504U5, Q5VUM8 | Gene names | SHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein B. | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.022613 (rank : 135) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.022604 (rank : 136) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
YTHD1_HUMAN
|
||||||
| NC score | 0.022511 (rank : 137) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
MUC15_MOUSE
|
||||||
| NC score | 0.022472 (rank : 138) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C6Z1, Q3UQC4, Q6XYQ9, Q7TMH6, Q8CE82 | Gene names | Muc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.022301 (rank : 139) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.022283 (rank : 140) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
K2002_HUMAN
|
||||||
| NC score | 0.021901 (rank : 141) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H792, Q6ZS78, Q8NCM3 | Gene names | KIAA2002 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized tyrosine-protein kinase KIAA2002 (EC 2.7.10.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.021779 (rank : 142) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.021570 (rank : 143) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.021025 (rank : 144) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.020264 (rank : 145) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
I12R2_HUMAN
|
||||||
| NC score | 0.018627 (rank : 146) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99665 | Gene names | IL12RB2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-2 chain precursor (IL-12 receptor beta-2) (IL-12R-beta2). | |||||
|
CENPU_HUMAN
|
||||||
| NC score | 0.018437 (rank : 147) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.018215 (rank : 148) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.017213 (rank : 149) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.016646 (rank : 150) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.016481 (rank : 151) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.016315 (rank : 152) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.016271 (rank : 153) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.015826 (rank : 154) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.015769 (rank : 155) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
LSP1_MOUSE
|
||||||
| NC score | 0.015274 (rank : 156) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19973, P97339, Q04950, Q62022, Q62023, Q62024, Q8CD28, Q99L65 | Gene names | Lsp1, Pp52, S37, Wp34 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (S37 protein). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.014500 (rank : 157) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
PHAR3_HUMAN
|
||||||
| NC score | 0.013990 (rank : 158) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96KR7, Q707P6, Q9H4T4 | Gene names | PHACTR3, C20orf101, SCAPIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.013101 (rank : 159) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.013039 (rank : 160) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
LAMB3_MOUSE
|
||||||
| NC score | 0.012325 (rank : 161) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.012264 (rank : 162) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.011930 (rank : 163) | θ value | 8.99809 (rank : 187) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
HRSL5_HUMAN
|
||||||
| NC score | 0.011435 (rank : 164) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96KN8 | Gene names | HRASLS5, HRLP5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.011333 (rank : 165) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.011149 (rank : 166) | θ value | 8.99809 (rank : 186) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.011131 (rank : 167) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.010318 (rank : 168) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.010281 (rank : 169) | θ value | 8.99809 (rank : 185) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
FBXL5_MOUSE
|
||||||
| NC score | 0.008956 (rank : 170) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C2S5, Q3TWC9, Q80XI5, Q8BGF5, Q8BNL3, Q8C3Q8 | Gene names | Fbxl5 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 5 (F-box and leucine-rich repeat protein 5). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.008749 (rank : 171) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| NC score | 0.008565 (rank : 172) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.008518 (rank : 173) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.008228 (rank : 174) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
ST2B1_HUMAN
|
||||||
| NC score | 0.008139 (rank : 175) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00204, O00205, O75814 | Gene names | SULT2B1, HSST2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase family cytosolic 2B member 1 (EC 2.8.2.2) (Sulfotransferase 2B1) (Alcohol sulfotransferase) (Hydroxysteroid sulfotransferase 2). | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.007996 (rank : 176) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
GBX1_HUMAN
|
||||||
| NC score | 0.007956 (rank : 177) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14549 | Gene names | GBX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein GBX-1 (Gastrulation and brain-specific homeobox protein 1). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.007955 (rank : 178) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
SOX3_HUMAN
|
||||||
| NC score | 0.007135 (rank : 179) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41225, P35714, Q9NP49 | Gene names | SOX3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-3. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.006941 (rank : 180) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.006389 (rank : 181) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
IRAK1_MOUSE
|
||||||
| NC score | 0.005691 (rank : 182) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62406, Q6Y3Z5, Q6Y3Z6 | Gene names | Irak1, Il1rak | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor-associated kinase 1 (EC 2.7.11.1) (IRAK-1) (IRAK) (Pelle-like protein kinase) (mPLK). | |||||
|
ADA12_HUMAN
|
||||||
| NC score | 0.004766 (rank : 183) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43184, O60470 | Gene names | ADAM12, MLTN | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.004396 (rank : 184) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
FOXF2_HUMAN
|
||||||
| NC score | 0.004050 (rank : 185) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12947, Q9UQ85 | Gene names | FOXF2, FKHL6, FREAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F2 (Forkhead-related protein FKHL6) (Forkhead- related transcription factor 2) (FREAC-2) (Forkhead-related activator 2). | |||||
|
FOXGB_HUMAN
|
||||||
| NC score | 0.003921 (rank : 186) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
PGLB_HUMAN
|
||||||
| NC score | 0.003730 (rank : 187) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99645, Q8NEJ5 | Gene names | DSPG3, PGLB | |||
|
Domain Architecture |

|
|||||
| Description | Dermatan sulfate proteoglycan 3 precursor (Epiphycan) (Small chondroitin/dermatan sulfate proteoglycan) (Proteoglycan-Lb) (PG-Lb). | |||||
|
SBK1_MOUSE
|
||||||
| NC score | 0.002865 (rank : 188) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8QZX0 | Gene names | Sbk1, Sbk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SBK1 (EC 2.7.11.1) (SH3-binding kinase 1). | |||||
|
PTPRJ_HUMAN
|
||||||
| NC score | 0.002580 (rank : 189) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12913, Q15255, Q8NHM2 | Gene names | PTPRJ, DEP1 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase eta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase eta) (R-PTP-eta) (HPTP eta) (Protein- tyrosine phosphatase receptor type J) (Density-enhanced phosphatase 1) (DEP-1) (CD148 antigen). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.002532 (rank : 190) | θ value | 1.06291 (rank : 73) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.001061 (rank : 191) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
EGR4_MOUSE
|
||||||
| NC score | -0.000146 (rank : 192) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
TIE1_HUMAN
|
||||||
| NC score | -0.000265 (rank : 193) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | -0.000309 (rank : 194) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
KLF3_MOUSE
|
||||||
| NC score | -0.000505 (rank : 195) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60980 | Gene names | Klf3, Bklf | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 3 (Basic krueppel-like factor) (CACCC-box-binding protein BKLF) (TEF-2). | |||||
|
SCRT1_MOUSE
|
||||||
| NC score | -0.000749 (rank : 196) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 188 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99M85 | Gene names | Scrt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor scratch 1 (Scratch homolog 1 zinc finger protein) (Scratch 1) (SCRT) (mScrt). | |||||