Please be patient as the page loads
|
UBE4B_HUMAN
|
||||||
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UBE4B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
UBE4B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.969984 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
UBE4A_HUMAN
|
||||||
| θ value | 6.31527e-70 (rank : 3) | NC score | 0.806304 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14139, Q6P5T4, Q7Z639 | Gene names | UBE4A, KIAA0126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin conjugation factor E4 A. | |||||
|
RNF37_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 4) | NC score | 0.233299 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q925F4 | Gene names | Ubox5, Rnf37, Ubce7ip5, Uip5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (UbcM4-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
RNF37_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 5) | NC score | 0.198902 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94941 | Gene names | UBOX5, KIAA0860, RNF37, UBCE7IP5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.140014 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.142445 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.097302 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.083102 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.057152 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.068304 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
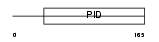
|
|||||
| Description | Disabled homolog 1. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.078155 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.052953 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
FOXK1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.027009 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |
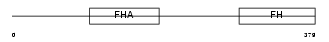
|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.084358 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
STK6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.004502 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
KCD18_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.055786 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.041514 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.066478 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.041660 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.014579 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.058931 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
NDC1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.069410 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCB1 | Gene names | Tmem48, Ndc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NDC1 (Transmembrane protein 48). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.069678 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MGRN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.046581 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
ADCY3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.037097 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.061886 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
LRCH4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.024656 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.083305 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
RBMS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.033434 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
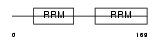
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
RBMS1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.033329 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.077221 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.033612 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.039000 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.046208 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRCH4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.023963 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.052943 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.062672 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
NDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.062080 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTX1, Q8NB76, Q9H9T6, Q9NSG3, Q9NSG4, Q9NVZ7 | Gene names | TMEM48, NDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NDC1 (hNDC1) (Transmembrane protein 48). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.034760 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PER2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.037708 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.045111 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.080875 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.055174 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.015386 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
ADCY3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.031968 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.080507 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.067030 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO039_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.053476 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.016852 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EPN2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.032106 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.053266 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.020513 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
TENS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.046239 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
TTC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.044425 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.021211 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
ENH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.019880 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9N2J9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-H_3q26 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (Env protein HERV-H/p60) (HERV-H/env60) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
HSF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.046458 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
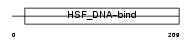
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.051882 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
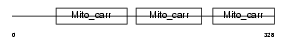
MPCP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.033108 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEM8, Q542V7 | Gene names | Slc25a3 | |||
|
Domain Architecture |
|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.045319 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.014899 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.034485 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
OTP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.010077 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09113 | Gene names | Otp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein orthopedia. | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.058575 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ZN275_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.000616 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSD4 | Gene names | ZNF275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 275. | |||||
|
CBL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.035988 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.056544 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.029730 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.040586 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
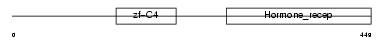
RXRA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.007476 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
VAX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.008320 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SQQ9, Q6ZSX0 | Gene names | VAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
COE3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.019099 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
DOS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.057995 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.033839 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.038118 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
NUFP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.028524 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PROP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.011004 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.036148 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.039315 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.024626 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.032006 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.015960 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.057075 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.004505 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.045699 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
COE3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.018287 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
CP4Z1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.009146 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W10 | Gene names | CYP4Z1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4Z1 (EC 1.14.14.1) (CYPIVZ1). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.013885 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.049328 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.020911 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
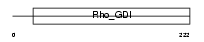
GDIT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.027845 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62160 | Gene names | Arhgdig, Gdi5 | |||
|
Domain Architecture |
|
|||||
| Description | Rho GDP-dissociation inhibitor 3 (Rho GDI 3) (Rho-GDI gamma) (Rho- GDI2). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.073039 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
K0460_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.035692 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.038657 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NRIP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.022311 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.030011 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PHAR3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.021556 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYK5, Q8BYS8, Q8C058, Q9DB87 | Gene names | Phactr3, Scapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
PRR8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.044429 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.049318 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.022522 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.025135 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
STXB5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.023162 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T5C0, Q5T5C1, Q5T5C2, Q8NBG8, Q96NG9 | Gene names | STXBP5, LLGL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
STXB5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.023335 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K400, Q80TM5, Q8K401, Q9D2F3 | Gene names | Stxbp5, Kiaa4253, Llgl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.051631 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TBC19_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.053307 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5T2 | Gene names | TBC1D19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 19. | |||||
|
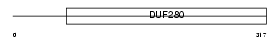
ADMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.003541 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 633 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15218 | Gene names | ADMR | |||
|
Domain Architecture |
|
|||||
| Description | Adrenomedullin receptor (AM-R). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.017973 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.043260 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CDKL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.002413 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.051447 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.012323 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
E2F4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.022283 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
EPN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.036707 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.030993 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.015669 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.016496 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.052215 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.029965 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.026475 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.051886 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
ZFP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | -0.000455 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NJ6 | Gene names | ZFP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 3 homolog (Zfp-3). | |||||
|
ADA19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.011175 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.004843 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.036724 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.010324 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.021775 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HIPK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.002519 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2X6, Q75MR7, Q8WWI4, Q9H2Y1 | Gene names | HIPK2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (hHIPk2). | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.018976 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.038419 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.019907 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18428, O43438, Q92672, Q9H403, Q9UD66 | Gene names | LBP | |||
|
Domain Architecture |
|
|||||
| Description | Lipopolysaccharide-binding protein precursor (LBP). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.012626 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.008364 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.008487 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.008227 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.008492 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.005333 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PAX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.011046 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02548, O75933 | Gene names | PAX5 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PAX5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.011167 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02650 | Gene names | Pax5, Pax-5 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.006663 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.033026 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.032258 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TF3B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.017259 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92994, Q13223, Q5PR24, Q6IQ02, Q9HCW6, Q9HCW7, Q9HCW8 | Gene names | BRF1, BRF | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor IIIB 90 kDa subunit (TFIIIB90) (hTFIIIB90) (B- related factor 1) (BRF-1) (hBRF) (TATA box-binding protein-associated factor, RNA polymerase III, subunit 2) (TAF3B2). | |||||
|
ZBT45_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.003679 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
ATS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.005643 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.039383 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.013675 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CEP68_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.015627 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
COMD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.024371 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0A8, Q7L637, Q9NX43 | Gene names | COMMD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COMM domain-containing protein 4. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.026455 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.021584 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.031228 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
LRC56_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.014270 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.019301 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.041310 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.041486 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.029245 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.038670 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
BSN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.051056 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.051436 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.056384 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.076565 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NU214_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050166 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.055318 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.061017 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.051039 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.055210 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.054420 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
UBE4B_MOUSE
|
||||||
| NC score | 0.969984 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
UBE4A_HUMAN
|
||||||
| NC score | 0.806304 (rank : 3) | θ value | 6.31527e-70 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14139, Q6P5T4, Q7Z639 | Gene names | UBE4A, KIAA0126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin conjugation factor E4 A. | |||||
|
RNF37_MOUSE
|
||||||
| NC score | 0.233299 (rank : 4) | θ value | 0.00020696 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q925F4 | Gene names | Ubox5, Rnf37, Ubce7ip5, Uip5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (UbcM4-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
RNF37_HUMAN
|
||||||
| NC score | 0.198902 (rank : 5) | θ value | 0.00665767 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O94941 | Gene names | UBOX5, KIAA0860, RNF37, UBCE7IP5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.142445 (rank : 6) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.140014 (rank : 7) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.097302 (rank : 8) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.084358 (rank : 9) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.083305 (rank : 10) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.083102 (rank : 11) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.080875 (rank : 12) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.080507 (rank : 13) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.078155 (rank : 14) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.077221 (rank : 15) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.076565 (rank : 16) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.073039 (rank : 17) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.069678 (rank : 18) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NDC1_MOUSE
|
||||||
| NC score | 0.069410 (rank : 19) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCB1 | Gene names | Tmem48, Ndc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NDC1 (Transmembrane protein 48). | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.068304 (rank : 20) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.067030 (rank : 21) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.066478 (rank : 22) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.062672 (rank : 23) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
NDC1_HUMAN
|
||||||
| NC score | 0.062080 (rank : 24) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BTX1, Q8NB76, Q9H9T6, Q9NSG3, Q9NSG4, Q9NVZ7 | Gene names | TMEM48, NDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin NDC1 (hNDC1) (Transmembrane protein 48). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.061886 (rank : 25) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.061017 (rank : 26) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.058931 (rank : 27) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.058575 (rank : 28) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
DOS_HUMAN
|
||||||
| NC score | 0.057995 (rank : 29) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N350, O43385 | Gene names | DOS, C19orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Dos. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.057152 (rank : 30) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.057075 (rank : 31) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.056544 (rank : 32) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.056384 (rank : 33) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
KCD18_HUMAN
|
||||||
| NC score | 0.055786 (rank : 34) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.055318 (rank : 35) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.055210 (rank : 36) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.055174 (rank : 37) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.054420 (rank : 38) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CO039_HUMAN
|
||||||
| NC score | 0.053476 (rank : 39) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6ZRI6, Q71JB1, Q7L3S0, Q8N3F2, Q96FB6, Q9NTU5 | Gene names | C15orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39. | |||||
|
TBC19_HUMAN
|
||||||
| NC score | 0.053307 (rank : 40) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N5T2 | Gene names | TBC1D19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 19. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.053266 (rank : 41) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.052953 (rank : 42) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.052943 (rank : 43) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.052215 (rank : 44) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.051886 (rank : 45) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.051882 (rank : 46) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.051631 (rank : 47) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.051447 (rank : 48) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.051436 (rank : 49) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.051056 (rank : 50) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.051039 (rank : 51) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.050166 (rank : 52) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.049328 (rank : 53) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.049318 (rank : 54) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MGRN1_HUMAN
|
||||||
| NC score | 0.046581 (rank : 55) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60291, Q86W76 | Gene names | MGRN1, KIAA0544, RNF156 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase MGRN1 (EC 6.3.2.-) (Mahogunin ring finger protein 1) (RING finger protein 156). | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.046458 (rank : 56) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
TENS1_HUMAN
|
||||||
| NC score | 0.046239 (rank : 57) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBL0 | Gene names | TNS1, TNS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.046208 (rank : 58) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.045699 (rank : 59) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.045319 (rank : 60) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.045111 (rank : 61) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.044429 (rank : 62) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
TTC1_MOUSE
|
||||||
| NC score | 0.044425 (rank : 63) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91Z38, Q3TLM0 | Gene names | Ttc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 1 (TPR repeat protein 1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.043260 (rank : 64) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.041660 (rank : 65) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.041514 (rank : 66) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.041486 (rank : 67) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.041310 (rank : 68) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.040586 (rank : 69) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.039383 (rank : 70) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.039315 (rank : 71) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.039000 (rank : 72) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.038670 (rank : 73) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.038657 (rank : 74) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.038419 (rank : 75) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.038118 (rank : 76) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.037708 (rank : 77) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
ADCY3_MOUSE
|
||||||
| NC score | 0.037097 (rank : 78) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHH7 | Gene names | Adcy3 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.036724 (rank : 79) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
EPN1_HUMAN
|
||||||
| NC score | 0.036707 (rank : 80) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6I3, Q86ST3, Q9HA18 | Gene names | EPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (EH domain-binding mitotic phosphoprotein). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.036148 (rank : 81) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.035988 (rank : 82) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.035692 (rank : 83) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.034760 (rank : 84) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.034485 (rank : 85) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.033839 (rank : 86) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.033612 (rank : 87) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.033434 (rank : 88) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
RBMS1_MOUSE
|
||||||
| NC score | 0.033329 (rank : 89) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91W59, Q6PEU6, Q6PHC2, Q9WTK4 | Gene names | Rbms1, Mssp, Mssp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1). | |||||
|
MPCP_MOUSE
|
||||||
| NC score | 0.033108 (rank : 90) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEM8, Q542V7 | Gene names | Slc25a3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphate carrier protein, mitochondrial precursor (PTP) (Solute carrier family 25 member 3). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.033026 (rank : 91) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.032258 (rank : 92) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
EPN2_MOUSE
|
||||||
| NC score | 0.032106 (rank : 93) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHU3, O70447, Q8BZ85 | Gene names | Epn2 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2) (Intersectin-EH-binding protein 2) (Ibp2). | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.032006 (rank : 94) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
ADCY3_HUMAN
|
||||||
| NC score | 0.031968 (rank : 95) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60266, Q9UDB1 | Gene names | ADCY3, KIAA0511 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylate cyclase type 3 (EC 4.6.1.1) (Adenylate cyclase type III) (Adenylate cyclase, olfactive type) (ATP pyrophosphate-lyase 3) (Adenylyl cyclase 3) (AC-III) (AC3). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.031228 (rank : 96) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.030993 (rank : 97) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.030011 (rank : 98) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.029965 (rank : 99) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.029730 (rank : 100) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.029245 (rank : 101) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
NUFP1_MOUSE
|
||||||
| NC score | 0.028524 (rank : 102) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
GDIT_MOUSE
|
||||||
| NC score | 0.027845 (rank : 103) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62160 | Gene names | Arhgdig, Gdi5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho GDP-dissociation inhibitor 3 (Rho GDI 3) (Rho-GDI gamma) (Rho- GDI2). | |||||
|
FOXK1_MOUSE
|
||||||
| NC score | 0.027009 (rank : 104) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42128, O35939 | Gene names | Foxk1, Mnf | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K1 (Myocyte nuclear factor) (MNF). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.026475 (rank : 105) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.026455 (rank : 106) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.025135 (rank : 107) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
LRCH4_HUMAN
|
||||||
| NC score | 0.024656 (rank : 108) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.024626 (rank : 109) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
COMD4_HUMAN
|
||||||
| NC score | 0.024371 (rank : 110) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0A8, Q7L637, Q9NX43 | Gene names | COMMD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COMM domain-containing protein 4. | |||||
|
LRCH4_MOUSE
|
||||||
| NC score | 0.023963 (rank : 111) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
STXB5_MOUSE
|
||||||
| NC score | 0.023335 (rank : 112) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K400, Q80TM5, Q8K401, Q9D2F3 | Gene names | Stxbp5, Kiaa4253, Llgl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
STXB5_HUMAN
|
||||||
| NC score | 0.023162 (rank : 113) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T5C0, Q5T5C1, Q5T5C2, Q8NBG8, Q96NG9 | Gene names | STXBP5, LLGL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 5 (Tomosyn-1) (Lethal(2) giant larvae protein homolog 3). | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.022522 (rank : 114) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
NRIP1_MOUSE
|
||||||
| NC score | 0.022311 (rank : 115) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
E2F4_HUMAN
|
||||||
| NC score | 0.022283 (rank : 116) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.021775 (rank : 117) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.021584 (rank : 118) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PHAR3_MOUSE
|
||||||
| NC score | 0.021556 (rank : 119) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYK5, Q8BYS8, Q8C058, Q9DB87 | Gene names | Phactr3, Scapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.021211 (rank : 120) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.020911 (rank : 121) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.020513 (rank : 122) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
LBP_HUMAN
|
||||||
| NC score | 0.019907 (rank : 123) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18428, O43438, Q92672, Q9H403, Q9UD66 | Gene names | LBP | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-binding protein precursor (LBP). | |||||
|
ENH2_HUMAN
|
||||||
| NC score | 0.019880 (rank : 124) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9N2J9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HERV-H_3q26 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (Env protein HERV-H/p60) (HERV-H/env60) [Contains: Surface protein (SU); Transmembrane protein (TM)]. | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.019301 (rank : 125) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
COE3_HUMAN
|
||||||
| NC score | 0.019099 (rank : 126) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.018976 (rank : 127) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
COE3_MOUSE
|
||||||
| NC score | 0.018287 (rank : 128) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017973 (rank : 129) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TF3B_HUMAN
|
||||||
| NC score | 0.017259 (rank : 130) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92994, Q13223, Q5PR24, Q6IQ02, Q9HCW6, Q9HCW7, Q9HCW8 | Gene names | BRF1, BRF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor IIIB 90 kDa subunit (TFIIIB90) (hTFIIIB90) (B- related factor 1) (BRF-1) (hBRF) (TATA box-binding protein-associated factor, RNA polymerase III, subunit 2) (TAF3B2). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.016852 (rank : 131) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.016496 (rank : 132) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.015960 (rank : 133) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.015669 (rank : 134) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CEP68_MOUSE
|
||||||
| NC score | 0.015627 (rank : 135) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.015386 (rank : 136) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.014899 (rank : 137) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.014579 (rank : 138) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
LRC56_HUMAN
|
||||||
| NC score | 0.014270 (rank : 139) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IYG6, Q8N3Q4 | Gene names | LRRC56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 56. | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.013885 (rank : 140) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.013675 (rank : 141) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.012626 (rank : 142) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.012323 (rank : 143) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
ADA19_HUMAN
|
||||||
| NC score | 0.011175 (rank : 144) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H013, Q9BZL5, Q9UHP2 | Gene names | ADAM19, MLTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta) (Metalloprotease and disintegrin dentritic antigen marker) (MADDAM). | |||||
|
PAX5_MOUSE
|
||||||
| NC score | 0.011167 (rank : 145) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02650 | Gene names | Pax5, Pax-5 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PAX5_HUMAN
|
||||||
| NC score | 0.011046 (rank : 146) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02548, O75933 | Gene names | PAX5 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-5 (B-cell-specific transcription factor) (BSAP). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.011004 (rank : 147) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.010324 (rank : 148) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
OTP_MOUSE
|
||||||
| NC score | 0.010077 (rank : 149) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09113 | Gene names | Otp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein orthopedia. | |||||
|
CP4Z1_HUMAN
|
||||||
| NC score | 0.009146 (rank : 150) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W10 | Gene names | CYP4Z1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4Z1 (EC 1.14.14.1) (CYPIVZ1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.008492 (rank : 151) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.008487 (rank : 152) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.008364 (rank : 153) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
VAX1_HUMAN
|
||||||
| NC score | 0.008320 (rank : 154) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SQQ9, Q6ZSX0 | Gene names | VAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ventral anterior homeobox 1. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.008227 (rank : 155) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.007476 (rank : 156) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.006663 (rank : 157) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
ATS2_HUMAN
|
||||||
| NC score | 0.005643 (rank : 158) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95450 | Gene names | ADAMTS2, PCINP, PCPNI | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-2 precursor (EC 3.4.24.14) (A disintegrin and metalloproteinase with thrombospondin motifs 2) (ADAM-TS 2) (ADAM-TS2) (Procollagen I/II amino propeptide-processing enzyme) (Procollagen I N-proteinase) (PC I-NP) (Procollagen N-endopeptidase) (pNPI). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.005333 (rank : 159) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.004843 (rank : 160) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.004505 (rank : 161) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
STK6_MOUSE
|
||||||
| NC score | 0.004502 (rank : 162) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97477, O35624, Q91YU4 | Gene names | Stk6, Ark1, Aurka, Ayk1, Iak1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 6 (EC 2.7.11.1) (Aurora-family kinase 1) (Aurora/IPL1-related kinase 1) (Ipl1- and aurora-related kinase 1) (Aurora-A) (Serine/threonine-protein kinase Ayk1). | |||||
|
ZBT45_HUMAN
|
||||||
| NC score | 0.003679 (rank : 163) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96K62 | Gene names | ZBTB45, ZNF499 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 45 (Zinc finger protein 499). | |||||
|
ADMR_HUMAN
|
||||||
| NC score | 0.003541 (rank : 164) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 633 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15218 | Gene names | ADMR | |||
|
Domain Architecture |

|
|||||
| Description | Adrenomedullin receptor (AM-R). | |||||
|
HIPK2_HUMAN
|
||||||
| NC score | 0.002519 (rank : 165) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H2X6, Q75MR7, Q8WWI4, Q9H2Y1 | Gene names | HIPK2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (hHIPk2). | |||||
|
CDKL5_HUMAN
|
||||||
| NC score | 0.002413 (rank : 166) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O76039, Q14198, Q5H985, Q9UJL6 | Gene names | CDKL5, STK9 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase-like 5 (EC 2.7.11.22) (Serine/threonine- protein kinase 9). | |||||
|
ZN275_HUMAN
|
||||||
| NC score | 0.000616 (rank : 167) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NSD4 | Gene names | ZNF275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 275. | |||||
|
ZFP3_HUMAN
|
||||||
| NC score | -0.000455 (rank : 168) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96NJ6 | Gene names | ZFP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 3 homolog (Zfp-3). | |||||