Please be patient as the page loads
|
CEP68_MOUSE
|
||||||
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CEP68_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.932812 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
CEP68_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 3) | NC score | 0.353256 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 4) | NC score | 0.155730 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.048451 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.082981 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 7) | NC score | 0.092942 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.084337 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.059745 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
BNC1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.050972 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.040615 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TRI55_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.033715 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |
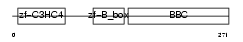
|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.076926 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.041578 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.026045 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.051928 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.037651 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.080021 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.034082 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.060642 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.024169 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.063025 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.029239 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.011603 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.038931 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.030476 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CRUM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.008335 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
KSYK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.004083 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
PAR10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.047453 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.040816 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
ZN384_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.002404 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF68, O15407, Q8N938 | Gene names | ZNF384, CAGH1, CIZ, NMP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 384 (Nuclear matrix transcription factor 4) (Nuclear matrix protein 4) (CAG repeat protein 1) (CAS-interacting zinc finger protein). | |||||
|
BMP6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.011434 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20722 | Gene names | Bmp6, Bmp-6, Vgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 6 precursor (BMP-6) (VG-1-related protein) (VGR-1). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.067465 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.033552 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
PAX3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.009817 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |
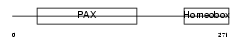
|
|||||
| Description | Paired box protein Pax-3. | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.003510 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.033027 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.018693 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.032144 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.014816 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
LPIN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.013838 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |
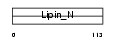
|
|||||
| Description | Lipin-3. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.035216 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
UT14A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.018913 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.065073 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.014304 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
CO027_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.021761 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2M3C6, Q8N993, Q96LL5 | Gene names | C15orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.019808 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.015722 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
POLS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.003225 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5K2P8 | Gene names | Prss36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.037025 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.016205 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
YETS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.025521 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ZN533_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.016839 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
B3GT4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.010506 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96024 | Gene names | B3GALT4, GALT4 | |||
|
Domain Architecture |
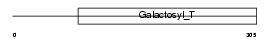
|
|||||
| Description | Beta-1,3-galactosyltransferase 4 (EC 2.4.1.62) (Beta-1,3-GalTase 4) (Beta3Gal-T4) (b3Gal-T4) (Ganglioside galactosyltransferase) (UDP- galactose:beta-N-acetyl-galactosamine-beta-1,3-galactosyltransferase) (GAL-T2) (GalT4). | |||||
|
CM007_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.014105 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.060111 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
PRLPM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.009616 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHK0, Q9JI05 | Gene names | Prlpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental prolactin-like protein M precursor (PRL-like protein M) (PLP-M). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.031485 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
SIRT6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.012245 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59941 | Gene names | Sirt6, Sir2l6 | |||
|
Domain Architecture |
|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.008026 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.015627 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
YTHD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.012781 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5A9, Q5VSZ9, Q8TDH0, Q9BUJ5 | Gene names | YTHDF2, HGRG8 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 2 (High-glucose-regulated protein 8) (NY- REN-2 antigen) (CLL-associated antigen KW-14). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.053557 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.050211 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.060860 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CEP68_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.932812 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.353256 (rank : 3) | θ value | 1.58096e-12 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.155730 (rank : 4) | θ value | 1.43324e-05 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.092942 (rank : 5) | θ value | 0.0330416 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.084337 (rank : 6) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.082981 (rank : 7) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.080021 (rank : 8) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.076926 (rank : 9) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.067465 (rank : 10) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.065073 (rank : 11) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.063025 (rank : 12) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.060860 (rank : 13) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.060642 (rank : 14) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.060111 (rank : 15) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.059745 (rank : 16) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.053557 (rank : 17) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.051928 (rank : 18) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
BNC1_HUMAN
|
||||||
| NC score | 0.050972 (rank : 19) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.050211 (rank : 20) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.048451 (rank : 21) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
PAR10_HUMAN
|
||||||
| NC score | 0.047453 (rank : 22) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53GL7, Q8N2I0, Q8WV05, Q96CH7 | Gene names | PARP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly [ADP-ribose] polymerase 10 (EC 2.4.2.30) (PARP-10). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.041578 (rank : 23) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.040816 (rank : 24) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.040615 (rank : 25) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.038931 (rank : 26) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.037651 (rank : 27) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.037025 (rank : 28) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.035216 (rank : 29) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.034082 (rank : 30) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
TRI55_HUMAN
|
||||||
| NC score | 0.033715 (rank : 31) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.033552 (rank : 32) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.033027 (rank : 33) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.032144 (rank : 34) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.031485 (rank : 35) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.030476 (rank : 36) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.029239 (rank : 37) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.026045 (rank : 38) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
YETS2_MOUSE
|
||||||
| NC score | 0.025521 (rank : 39) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3TUF7, Q6PGF8, Q80TI2, Q8CG86 | Gene names | Yeats2, Kiaa1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.024169 (rank : 40) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
CO027_HUMAN
|
||||||
| NC score | 0.021761 (rank : 41) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2M3C6, Q8N993, Q96LL5 | Gene names | C15orf27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf27. | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.019808 (rank : 42) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
UT14A_MOUSE
|
||||||
| NC score | 0.018913 (rank : 43) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q640M1, Q6A094, Q6EJB4, Q6P6L1, Q9CZK8 | Gene names | Utp14a, JsdX, Kiaa0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Juvenile spermatogonial depletion-like X-linked protein) (Jsd-like X-linked protein). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.018693 (rank : 44) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ZN533_MOUSE
|
||||||
| NC score | 0.016839 (rank : 45) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXJ8, Q8BWQ7 | Gene names | Znf533, Zfp533 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 533. | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.016205 (rank : 46) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.015722 (rank : 47) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.015627 (rank : 48) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.014816 (rank : 49) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.014304 (rank : 50) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.014105 (rank : 51) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
LPIN3_MOUSE
|
||||||
| NC score | 0.013838 (rank : 52) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-3. | |||||
|
YTHD2_HUMAN
|
||||||
| NC score | 0.012781 (rank : 53) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5A9, Q5VSZ9, Q8TDH0, Q9BUJ5 | Gene names | YTHDF2, HGRG8 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 2 (High-glucose-regulated protein 8) (NY- REN-2 antigen) (CLL-associated antigen KW-14). | |||||
|
SIRT6_MOUSE
|
||||||
| NC score | 0.012245 (rank : 54) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59941 | Gene names | Sirt6, Sir2l6 | |||
|
Domain Architecture |

|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
HXB3_MOUSE
|
||||||
| NC score | 0.011603 (rank : 55) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
BMP6_MOUSE
|
||||||
| NC score | 0.011434 (rank : 56) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20722 | Gene names | Bmp6, Bmp-6, Vgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 6 precursor (BMP-6) (VG-1-related protein) (VGR-1). | |||||
|
B3GT4_HUMAN
|
||||||
| NC score | 0.010506 (rank : 57) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O96024 | Gene names | B3GALT4, GALT4 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,3-galactosyltransferase 4 (EC 2.4.1.62) (Beta-1,3-GalTase 4) (Beta3Gal-T4) (b3Gal-T4) (Ganglioside galactosyltransferase) (UDP- galactose:beta-N-acetyl-galactosamine-beta-1,3-galactosyltransferase) (GAL-T2) (GalT4). | |||||
|
PAX3_MOUSE
|
||||||
| NC score | 0.009817 (rank : 58) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24610, Q3UFQ9, Q8BRE7, Q9CXI6 | Gene names | Pax3, Pax-3 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3. | |||||
|
PRLPM_MOUSE
|
||||||
| NC score | 0.009616 (rank : 59) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHK0, Q9JI05 | Gene names | Prlpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Placental prolactin-like protein M precursor (PRL-like protein M) (PLP-M). | |||||
|
CRUM2_HUMAN
|
||||||
| NC score | 0.008335 (rank : 60) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.008026 (rank : 61) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
KSYK_MOUSE
|
||||||
| NC score | 0.004083 (rank : 62) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | 0.003510 (rank : 63) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
POLS2_MOUSE
|
||||||
| NC score | 0.003225 (rank : 64) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5K2P8 | Gene names | Prss36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
ZN384_HUMAN
|
||||||
| NC score | 0.002404 (rank : 65) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TF68, O15407, Q8N938 | Gene names | ZNF384, CAGH1, CIZ, NMP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 384 (Nuclear matrix transcription factor 4) (Nuclear matrix protein 4) (CAG repeat protein 1) (CAS-interacting zinc finger protein). | |||||