Please be patient as the page loads
|
SSH2_HUMAN
|
||||||
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SSH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.962722 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.965866 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988966 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 2.98686e-120 (rank : 5) | NC score | 0.953326 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 3.77839e-115 (rank : 6) | NC score | 0.951040 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 7) | NC score | 0.794644 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 6.84181e-24 (rank : 8) | NC score | 0.794302 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS1_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 9) | NC score | 0.775955 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS1_MOUSE
|
||||||
| θ value | 3.39556e-23 (rank : 10) | NC score | 0.772974 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 11) | NC score | 0.798712 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 12) | NC score | 0.779936 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 13) | NC score | 0.780913 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 14) | NC score | 0.767747 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 15) | NC score | 0.787276 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 2.69047e-20 (rank : 16) | NC score | 0.766742 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 17) | NC score | 0.771665 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 18) | NC score | 0.770803 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 19) | NC score | 0.769793 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS14_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 20) | NC score | 0.785477 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS14_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 21) | NC score | 0.781965 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS19_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 22) | NC score | 0.813953 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 23) | NC score | 0.760642 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS19_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 24) | NC score | 0.813232 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 25) | NC score | 0.745087 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 26) | NC score | 0.751755 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
STYX_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 27) | NC score | 0.794266 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 28) | NC score | 0.793609 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS22_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 29) | NC score | 0.784112 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 30) | NC score | 0.783989 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 31) | NC score | 0.744894 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS15_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 32) | NC score | 0.767475 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS21_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 33) | NC score | 0.744891 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 34) | NC score | 0.741974 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS12_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 35) | NC score | 0.716282 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS12_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 36) | NC score | 0.710566 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
DUS13_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 37) | NC score | 0.718086 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
DUS13_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 38) | NC score | 0.723431 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS3_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 39) | NC score | 0.731070 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 40) | NC score | 0.730200 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
STYL1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 41) | NC score | 0.708181 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
PTPM1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 42) | NC score | 0.469181 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
EDEM3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 43) | NC score | 0.042511 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.034965 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PTPM1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 45) | NC score | 0.438424 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 46) | NC score | 0.015017 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
PRIC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.017977 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.022373 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
BNC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.022361 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35914, O54886 | Gene names | Bnc1, Bnc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
ARHGA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.015398 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.028937 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.025191 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.253022 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.019723 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.031555 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
OXR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.019057 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.018984 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BNC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.024086 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
CEP68_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.024169 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
LPIN3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.014981 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.033412 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
RT18C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.031202 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3D5 | Gene names | MRPS18C | |||
|
Domain Architecture |
|
|||||
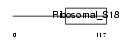
| Description | 28S ribosomal protein S18c, mitochondrial precursor (MRP-S18-c) (Mrps18c) (MRP-S18-1). | |||||
|
TEX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.023139 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.016855 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CI039_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.005357 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
MICA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.011551 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.013884 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.016901 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
SRPK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.006017 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.022284 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.005254 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ZN644_HUMAN
|
||||||
| θ value | 1.06291 (rank : 72) | NC score | 0.002713 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H582, Q5TCC0, Q6BEP7, Q6P446, Q6PI06, Q7LG67, Q9ULJ9 | Gene names | ZNF644, KIAA1221, ZEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 644 (Zinc finger motif enhancer-binding protein 2) (Zep-2). | |||||
|
ERRFI_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.029186 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
ERRFI_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.028507 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.012985 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CC14A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.192069 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CS021_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.019262 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVT2 | Gene names | C19orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21. | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.013847 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
EPM2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.238581 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
GLPT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.025293 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P57057, Q9HAQ1 | Gene names | SLC37A1, G3PP | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate transporter (G-3-P transporter) (G-3-P permease) (Solute carrier family 37 member 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.008979 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.007477 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.008376 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.008174 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.009211 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.018801 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.020788 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.001252 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
IKKA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | -0.002029 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15111, O14666, Q13132, Q92467 | Gene names | CHUK, IKKA | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase alpha subunit (EC 2.7.11.10) (I kappa-B kinase alpha) (IkBKA) (IKK-alpha) (IKK-A) (IkappaB kinase) (I-kappa-B kinase 1) (IKK1) (Conserved helix-loop- helix ubiquitous kinase) (Nuclear factor NF-kappa-B inhibitor kinase alpha) (NFKBIKA). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.002969 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.009843 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PRIC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.013596 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
ZN592_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.000585 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.031035 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.182057 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.206012 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.008605 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
IRX4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.006620 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY61 | Gene names | Irx4, Irxa3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-4 (Iroquois homeobox protein 4) (Homeodomain protein IRXA3). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.014849 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.012235 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.025999 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AF9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.018609 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
ANR50_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.000100 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.007409 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
EPMIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.025020 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEH5, Q80TS4 | Gene names | Epm2aip1, Kiaa0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
GBRT_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.002550 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLF1 | Gene names | Gabrq | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit theta precursor (GABA(A) receptor subunit theta). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.002521 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.009714 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.007244 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RELN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.008851 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
|
|||||
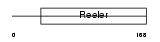
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
CC023_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.011937 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N3R3, Q495R1, Q495R3, Q6GMU8 | Gene names | C3orf23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C3orf23. | |||||
|
CX021_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.020037 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf21 homolog. | |||||
|
DUS23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.198680 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
EDRF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.010496 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
EPMIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.023182 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L775, O94866, Q9H3L3 | Gene names | EPM2AIP1, KIAA0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
ESCO2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.010476 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
FETUA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.012352 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29699, O35634 | Gene names | Ahsg, Fetua | |||
|
Domain Architecture |
|
|||||
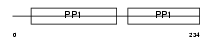
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Countertrypin). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.016680 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.004628 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.012153 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.005979 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.012162 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.016792 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
DUS23_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.178428 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.012770 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.016695 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PLK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | -0.004294 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.009177 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TENA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | -0.001277 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.010020 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
BCAR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.003672 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75815, Q6UW40, Q9BR50 | Gene names | BCAR3, NSP2, SH2D3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (SH2 domain- containing protein 3B) (Novel SH2-containing protein 2). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.009687 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CJ022_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.009312 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PDY2, Q3U2E1 | Gene names | Gm237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf22 homolog. | |||||
|
CQ028_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.008091 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
E2AK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | -0.003083 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
GNPTA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.006481 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3T906, Q3ZQK2, Q6IPW5, Q86TQ2, Q96N13, Q9ULL2 | Gene names | GNPTAB, GNPTA, KIAA1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
IKKA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | -0.002977 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60680, Q80VU2, Q9D2X3 | Gene names | Chuk, Ikka | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase alpha subunit (EC 2.7.11.10) (I kappa-B kinase alpha) (IkBKA) (IKK-alpha) (IKK-A) (IkappaB kinase) (I-kappa-B kinase 1) (IKK1) (Conserved helix-loop- helix ubiquitous kinase) (Nuclear factor NF-kappa-B inhibitor kinase alpha) (NFKBIKA). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.007535 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
STAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010581 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
ZN592_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | -0.000827 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92610 | Gene names | ZNF592, KIAA0211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592. | |||||
|
CC14B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.165705 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.050382 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
TP4A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.056593 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.054145 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.988966 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.965866 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.962722 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.953326 (rank : 5) | θ value | 2.98686e-120 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.951040 (rank : 6) | θ value | 3.77839e-115 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS19_MOUSE
|
||||||
| NC score | 0.813953 (rank : 7) | θ value | 2.5182e-18 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS19_HUMAN
|
||||||
| NC score | 0.813232 (rank : 8) | θ value | 7.32683e-18 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.798712 (rank : 9) | θ value | 3.75424e-22 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.794644 (rank : 10) | θ value | 3.07116e-24 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.794302 (rank : 11) | θ value | 6.84181e-24 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
STYX_MOUSE
|
||||||
| NC score | 0.794266 (rank : 12) | θ value | 6.20254e-17 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| NC score | 0.793609 (rank : 13) | θ value | 1.058e-16 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.787276 (rank : 14) | θ value | 1.5773e-20 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS14_MOUSE
|
||||||
| NC score | 0.785477 (rank : 15) | θ value | 5.07402e-19 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS22_MOUSE
|
||||||
| NC score | 0.784112 (rank : 16) | θ value | 6.41864e-14 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| NC score | 0.783989 (rank : 17) | θ value | 8.38298e-14 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS14_HUMAN
|
||||||
| NC score | 0.781965 (rank : 18) | θ value | 2.5182e-18 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.780913 (rank : 19) | θ value | 7.0802e-21 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.779936 (rank : 20) | θ value | 7.0802e-21 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS1_HUMAN
|
||||||
| NC score | 0.775955 (rank : 21) | θ value | 3.39556e-23 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS1_MOUSE
|
||||||
| NC score | 0.772974 (rank : 22) | θ value | 3.39556e-23 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.771665 (rank : 23) | θ value | 5.99374e-20 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.770803 (rank : 24) | θ value | 1.33526e-19 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.769793 (rank : 25) | θ value | 1.33526e-19 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.767747 (rank : 26) | θ value | 9.24701e-21 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS15_HUMAN
|
||||||
| NC score | 0.767475 (rank : 27) | θ value | 2.28291e-11 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.766742 (rank : 28) | θ value | 2.69047e-20 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.760642 (rank : 29) | θ value | 4.29542e-18 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.751755 (rank : 30) | θ value | 1.24977e-17 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.745087 (rank : 31) | θ value | 1.24977e-17 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.744894 (rank : 32) | θ value | 1.74796e-11 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS21_HUMAN
|
||||||
| NC score | 0.744891 (rank : 33) | θ value | 3.89403e-11 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_HUMAN
|
||||||
| NC score | 0.741974 (rank : 34) | θ value | 1.133e-10 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS3_MOUSE
|
||||||
| NC score | 0.731070 (rank : 35) | θ value | 1.06045e-08 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.730200 (rank : 36) | θ value | 5.26297e-08 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS13_MOUSE
|
||||||
| NC score | 0.723431 (rank : 37) | θ value | 8.11959e-09 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS13_HUMAN
|
||||||
| NC score | 0.718086 (rank : 38) | θ value | 3.64472e-09 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
DUS12_MOUSE
|
||||||
| NC score | 0.716282 (rank : 39) | θ value | 4.30538e-10 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS12_HUMAN
|
||||||
| NC score | 0.710566 (rank : 40) | θ value | 1.25267e-09 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
STYL1_HUMAN
|
||||||
| NC score | 0.708181 (rank : 41) | θ value | 4.45536e-07 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
PTPM1_MOUSE
|
||||||
| NC score | 0.469181 (rank : 42) | θ value | 0.00020696 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_HUMAN
|
||||||
| NC score | 0.438424 (rank : 43) | θ value | 0.0148317 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.253022 (rank : 44) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_HUMAN
|
||||||
| NC score | 0.238581 (rank : 45) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.206012 (rank : 46) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS23_HUMAN
|
||||||
| NC score | 0.198680 (rank : 47) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
CC14A_MOUSE
|
||||||
| NC score | 0.192069 (rank : 48) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.182057 (rank : 49) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
DUS23_MOUSE
|
||||||
| NC score | 0.178428 (rank : 50) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
CC14B_MOUSE
|
||||||
| NC score | 0.165705 (rank : 51) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
TP4A3_HUMAN
|
||||||
| NC score | 0.056593 (rank : 52) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| NC score | 0.054145 (rank : 53) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
DUS11_MOUSE
|
||||||
| NC score | 0.050382 (rank : 54) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.042511 (rank : 55) | θ value | 0.00102713 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.034965 (rank : 56) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.033412 (rank : 57) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.031555 (rank : 58) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
RT18C_HUMAN
|
||||||
| NC score | 0.031202 (rank : 59) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3D5 | Gene names | MRPS18C | |||
|
Domain Architecture |

|
|||||
| Description | 28S ribosomal protein S18c, mitochondrial precursor (MRP-S18-c) (Mrps18c) (MRP-S18-1). | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.031035 (rank : 60) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.029186 (rank : 61) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.028937 (rank : 62) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ERRFI_MOUSE
|
||||||
| NC score | 0.028507 (rank : 63) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JZ7 | Gene names | Errfi1, Mig6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein homolog) (Mig-6). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.025999 (rank : 64) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
GLPT_HUMAN
|
||||||
| NC score | 0.025293 (rank : 65) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P57057, Q9HAQ1 | Gene names | SLC37A1, G3PP | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate transporter (G-3-P transporter) (G-3-P permease) (Solute carrier family 37 member 1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.025191 (rank : 66) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EPMIP_MOUSE
|
||||||
| NC score | 0.025020 (rank : 67) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEH5, Q80TS4 | Gene names | Epm2aip1, Kiaa0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
CEP68_MOUSE
|
||||||
| NC score | 0.024169 (rank : 68) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0D9, Q3US49, Q5SW85, Q8R2V0 | Gene names | Cep68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
BNC1_HUMAN
|
||||||
| NC score | 0.024086 (rank : 69) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01954, Q15840 | Gene names | BNC1, BNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
EPMIP_HUMAN
|
||||||
| NC score | 0.023182 (rank : 70) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7L775, O94866, Q9H3L3 | Gene names | EPM2AIP1, KIAA0766 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EPM2A-interacting protein 1 (Laforin-interacting protein). | |||||
|
TEX2_MOUSE
|
||||||
| NC score | 0.023139 (rank : 71) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZPJ0 | Gene names | Tex2, Kiaa1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.022373 (rank : 72) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
BNC1_MOUSE
|
||||||
| NC score | 0.022361 (rank : 73) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35914, O54886 | Gene names | Bnc1, Bnc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-1. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.022284 (rank : 74) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.020788 (rank : 75) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
CX021_MOUSE
|
||||||
| NC score | 0.020037 (rank : 76) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf21 homolog. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.019723 (rank : 77) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CS021_HUMAN
|
||||||
| NC score | 0.019262 (rank : 78) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IVT2 | Gene names | C19orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21. | |||||
|
OXR1_HUMAN
|
||||||
| NC score | 0.019057 (rank : 79) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N573, Q3LIB5, Q6ZVK9, Q8N8V0, Q9H266, Q9NWC7 | Gene names | OXR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.018984 (rank : 80) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.018801 (rank : 81) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.018609 (rank : 82) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
PRIC2_HUMAN
|
||||||
| NC score | 0.017977 (rank : 83) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.016901 (rank : 84) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.016855 (rank : 85) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.016792 (rank : 86) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.016695 (rank : 87) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.016680 (rank : 88) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ARHGA_HUMAN
|
||||||
| NC score | 0.015398 (rank : 89) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15013, O14665, Q68D55, Q8IWD9, Q8IY77 | Gene names | ARHGEF10, KIAA0294 | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 10. | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.015017 (rank : 90) | θ value | 0.0563607 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
LPIN3_HUMAN
|
||||||
| NC score | 0.014981 (rank : 91) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQK8, Q5TDB9, Q9NPY8, Q9UJE5 | Gene names | LPIN3, LIPN3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-3 (Lipin 3-like). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.014849 (rank : 92) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.013884 (rank : 93) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.013847 (rank : 94) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
PRIC2_MOUSE
|
||||||
| NC score | 0.013596 (rank : 95) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.012985 (rank : 96) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.012770 (rank : 97) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
FETUA_MOUSE
|
||||||
| NC score | 0.012352 (rank : 98) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29699, O35634 | Gene names | Ahsg, Fetua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2-HS-glycoprotein precursor (Fetuin-A) (Countertrypin). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.012235 (rank : 99) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.012162 (rank : 100) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.012153 (rank : 101) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CC023_HUMAN
|
||||||
| NC score | 0.011937 (rank : 102) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N3R3, Q495R1, Q495R3, Q6GMU8 | Gene names | C3orf23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C3orf23. | |||||
|
MICA1_HUMAN
|
||||||
| NC score | 0.011551 (rank : 103) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
STAP2_MOUSE
|
||||||
| NC score | 0.010581 (rank : 104) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
EDRF1_MOUSE
|
||||||
| NC score | 0.010496 (rank : 105) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6GQV7, Q80ZY3, Q8R3P5 | Gene names | Edrf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Erythroid differentiation-related factor 1. | |||||
|
ESCO2_HUMAN
|
||||||
| NC score | 0.010476 (rank : 106) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.010020 (rank : 107) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.009843 (rank : 108) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.009714 (rank : 109) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.009687 (rank : 110) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CJ022_MOUSE
|
||||||
| NC score | 0.009312 (rank : 111) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PDY2, Q3U2E1 | Gene names | Gm237 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf22 homolog. | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.009211 (rank : 112) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.009177 (rank : 113) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.008979 (rank : 114) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.008851 (rank : 115) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.008605 (rank : 116) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.008376 (rank : 117) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.008174 (rank : 118) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
CQ028_HUMAN
|
||||||
| NC score | 0.008091 (rank : 119) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IV36, Q8N5L6, Q8TE83, Q9NT34 | Gene names | C17orf28, DMC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C17orf28 (Down-regulated in multiple cancers- 1). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.007535 (rank : 120) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.007477 (rank : 121) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.007409 (rank : 122) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.007244 (rank : 123) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
IRX4_MOUSE
|
||||||
| NC score | 0.006620 (rank : 124) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QY61 | Gene names | Irx4, Irxa3 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-4 (Iroquois homeobox protein 4) (Homeodomain protein IRXA3). | |||||
|
GNPTA_HUMAN
|
||||||
| NC score | 0.006481 (rank : 125) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3T906, Q3ZQK2, Q6IPW5, Q86TQ2, Q96N13, Q9ULL2 | Gene names | GNPTAB, GNPTA, KIAA1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
SRPK1_MOUSE
|
||||||
| NC score | 0.006017 (rank : 126) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O70551, O70193, Q99JT3 | Gene names | Srpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.005979 (rank : 127) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.005357 (rank : 128) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.005254 (rank : 129) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.004628 (rank : 130) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
BCAR3_HUMAN
|
||||||
| NC score | 0.003672 (rank : 131) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75815, Q6UW40, Q9BR50 | Gene names | BCAR3, NSP2, SH2D3B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (SH2 domain- containing protein 3B) (Novel SH2-containing protein 2). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.002969 (rank : 132) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ZN644_HUMAN
|
||||||
| NC score | 0.002713 (rank : 133) | θ value | 1.06291 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H582, Q5TCC0, Q6BEP7, Q6P446, Q6PI06, Q7LG67, Q9ULJ9 | Gene names | ZNF644, KIAA1221, ZEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 644 (Zinc finger motif enhancer-binding protein 2) (Zep-2). | |||||
|
GBRT_MOUSE
|
||||||
| NC score | 0.002550 (rank : 134) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLF1 | Gene names | Gabrq | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric-acid receptor subunit theta precursor (GABA(A) receptor subunit theta). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.002521 (rank : 135) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.001252 (rank : 136) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ZN592_MOUSE
|
||||||
| NC score | 0.000585 (rank : 137) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.000100 (rank : 138) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ZN592_HUMAN
|
||||||
| NC score | -0.000827 (rank : 139) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92610 | Gene names | ZNF592, KIAA0211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592. | |||||
|
TENA_HUMAN
|
||||||
| NC score | -0.001277 (rank : 140) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
IKKA_HUMAN
|
||||||
| NC score | -0.002029 (rank : 141) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15111, O14666, Q13132, Q92467 | Gene names | CHUK, IKKA | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase alpha subunit (EC 2.7.11.10) (I kappa-B kinase alpha) (IkBKA) (IKK-alpha) (IKK-A) (IkappaB kinase) (I-kappa-B kinase 1) (IKK1) (Conserved helix-loop- helix ubiquitous kinase) (Nuclear factor NF-kappa-B inhibitor kinase alpha) (NFKBIKA). | |||||
|
IKKA_MOUSE
|
||||||
| NC score | -0.002977 (rank : 142) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60680, Q80VU2, Q9D2X3 | Gene names | Chuk, Ikka | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of nuclear factor kappa-B kinase alpha subunit (EC 2.7.11.10) (I kappa-B kinase alpha) (IkBKA) (IKK-alpha) (IKK-A) (IkappaB kinase) (I-kappa-B kinase 1) (IKK1) (Conserved helix-loop- helix ubiquitous kinase) (Nuclear factor NF-kappa-B inhibitor kinase alpha) (NFKBIKA). | |||||
|
E2AK4_MOUSE
|
||||||
| NC score | -0.003083 (rank : 143) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 984 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QZ05, Q6GQT4, Q6ZPT5, Q8C5S0, Q8CIF5, Q9CT30, Q9CUV9, Q9ESB6, Q9ESB7, Q9ESB8 | Gene names | Eif2ak4, Gcn2, Kiaa1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein) (mGCN2). | |||||
|
PLK3_HUMAN
|
||||||
| NC score | -0.004294 (rank : 144) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4B4, Q15767, Q96CV1 | Gene names | PLK3, CNK, FNK, PRK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PLK3 (EC 2.7.11.21) (Polo-like kinase 3) (PLK-3) (Cytokine-inducible serine/threonine-protein kinase) (FGF- inducible kinase) (Proliferation-related kinase). | |||||