Please be patient as the page loads
|
ESCO2_HUMAN
|
||||||
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ESCO2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
ESCO2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.973741 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIB9, Q3UMT6, Q6IQX5, Q8BNG9, Q8BQF8, Q9CRI8 | Gene names | Esco2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 1.54831e-84 (rank : 3) | NC score | 0.782385 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
ESCO1_MOUSE
|
||||||
| θ value | 1.89267e-82 (rank : 4) | NC score | 0.876655 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 5) | NC score | 0.090120 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.064175 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.051572 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.072890 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.048246 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.033431 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.019595 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.037780 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.021273 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
PTPRR_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.014131 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.032198 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
CIR_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.071802 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.015606 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.032765 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.021228 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
TBL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.020043 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4P3, Q9UQE2 | Gene names | TBL2, WBSCR13 | |||
|
Domain Architecture |
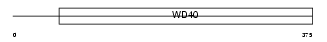
|
|||||
| Description | Transducin beta-like 2 protein (WS beta-transducin repeats protein) (WS-betaTRP) (Williams-Beuren syndrome chromosome region 13 protein). | |||||
|
CDA7L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.027031 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96GN5, Q6PIL4, Q86YT0, Q8IXN5, Q96C70, Q9H9A2, Q9NPV2 | Gene names | CDCA7L, HR1, JPO2, R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated 7-like protein (Protein JPO2) (Transcription factor RAM2). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.026429 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.027112 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.028081 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.030912 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.037579 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.014499 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.026765 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.009631 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
FA13C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.015429 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
MSH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.012262 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.008054 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.042548 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SEBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.019559 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.010476 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.007241 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.026327 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CG010_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.015323 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAC7, Q8TE00, Q8TEY1 | Gene names | C7orf10, DERP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C7orf10 (Dermal papilla-derived protein 13). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.058165 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.020626 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.038967 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.018053 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
ZN181_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | -0.000892 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2M3W8, Q49A75 | Gene names | ZNF181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 181 (HHZ181). | |||||
|
ATAD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.010815 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.009565 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.050478 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.060333 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.055694 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ESCO2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q56NI9 | Gene names | ESCO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
ESCO2_MOUSE
|
||||||
| NC score | 0.973741 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIB9, Q3UMT6, Q6IQX5, Q8BNG9, Q8BQF8, Q9CRI8 | Gene names | Esco2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO2 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 2) (ECO1 homolog 2). | |||||
|
ESCO1_MOUSE
|
||||||
| NC score | 0.876655 (rank : 3) | θ value | 1.89267e-82 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.782385 (rank : 4) | θ value | 1.54831e-84 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.090120 (rank : 5) | θ value | 9.29e-05 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.072890 (rank : 6) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.071802 (rank : 7) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.064175 (rank : 8) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.060333 (rank : 9) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.058165 (rank : 10) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.055694 (rank : 11) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.051572 (rank : 12) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.050478 (rank : 13) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.048246 (rank : 14) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.042548 (rank : 15) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.038967 (rank : 16) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.037780 (rank : 17) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.037579 (rank : 18) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.033431 (rank : 19) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.032765 (rank : 20) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.032198 (rank : 21) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.030912 (rank : 22) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.028081 (rank : 23) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.027112 (rank : 24) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CDA7L_HUMAN
|
||||||
| NC score | 0.027031 (rank : 25) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96GN5, Q6PIL4, Q86YT0, Q8IXN5, Q96C70, Q9H9A2, Q9NPV2 | Gene names | CDCA7L, HR1, JPO2, R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated 7-like protein (Protein JPO2) (Transcription factor RAM2). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.026765 (rank : 26) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.026429 (rank : 27) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.026327 (rank : 28) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.021273 (rank : 29) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.021228 (rank : 30) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.020626 (rank : 31) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
TBL2_HUMAN
|
||||||
| NC score | 0.020043 (rank : 32) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4P3, Q9UQE2 | Gene names | TBL2, WBSCR13 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin beta-like 2 protein (WS beta-transducin repeats protein) (WS-betaTRP) (Williams-Beuren syndrome chromosome region 13 protein). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.019595 (rank : 33) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
SEBP2_HUMAN
|
||||||
| NC score | 0.019559 (rank : 34) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.018053 (rank : 35) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.015606 (rank : 36) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
FA13C_HUMAN
|
||||||
| NC score | 0.015429 (rank : 37) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NE31, Q99787 | Gene names | FAM13C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
CG010_HUMAN
|
||||||
| NC score | 0.015323 (rank : 38) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAC7, Q8TE00, Q8TEY1 | Gene names | C7orf10, DERP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C7orf10 (Dermal papilla-derived protein 13). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.014499 (rank : 39) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
PTPRR_HUMAN
|
||||||
| NC score | 0.014131 (rank : 40) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15256, O00342, Q92682, Q9UE65 | Gene names | PTPRR, ECPTP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase R precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase PCPTP1) (NC-PTPCOM1) (Ch-1PTPase). | |||||
|
MSH3_HUMAN
|
||||||
| NC score | 0.012262 (rank : 41) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
ATAD2_HUMAN
|
||||||
| NC score | 0.010815 (rank : 42) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PL18, Q658P2, Q68CQ0, Q6PJV6, Q8N890, Q9UHS5 | Gene names | ATAD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 2. | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.010476 (rank : 43) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.009631 (rank : 44) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.009565 (rank : 45) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.008054 (rank : 46) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.007241 (rank : 47) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
ZN181_HUMAN
|
||||||
| NC score | -0.000892 (rank : 48) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q2M3W8, Q49A75 | Gene names | ZNF181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 181 (HHZ181). | |||||