Please be patient as the page loads
|
SSH1_HUMAN
|
||||||
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SSH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 164 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989644 (rank : 2) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.962722 (rank : 5) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.965279 (rank : 3) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 5.61992e-127 (rank : 5) | NC score | 0.963383 (rank : 4) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 3.52827e-121 (rank : 6) | NC score | 0.960700 (rank : 6) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 7) | NC score | 0.806299 (rank : 10) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 8) | NC score | 0.806252 (rank : 11) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 9) | NC score | 0.813717 (rank : 9) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS1_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 10) | NC score | 0.787749 (rank : 22) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS1_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 11) | NC score | 0.784427 (rank : 26) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 12) | NC score | 0.781771 (rank : 27) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 13) | NC score | 0.802236 (rank : 16) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 14) | NC score | 0.787240 (rank : 24) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 15) | NC score | 0.780882 (rank : 28) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 9.24701e-21 (rank : 16) | NC score | 0.787503 (rank : 23) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 17) | NC score | 0.794348 (rank : 20) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 18) | NC score | 0.795352 (rank : 19) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 19) | NC score | 0.785869 (rank : 25) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 20) | NC score | 0.762828 (rank : 31) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS19_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 21) | NC score | 0.830068 (rank : 7) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS14_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 22) | NC score | 0.796504 (rank : 18) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS14_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 23) | NC score | 0.799373 (rank : 17) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 24) | NC score | 0.767827 (rank : 30) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 25) | NC score | 0.775344 (rank : 29) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS19_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 26) | NC score | 0.828316 (rank : 8) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS22_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 27) | NC score | 0.805401 (rank : 14) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS22_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 28) | NC score | 0.805437 (rank : 13) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
STYX_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 29) | NC score | 0.806080 (rank : 12) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 30) | NC score | 0.804827 (rank : 15) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS15_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 31) | NC score | 0.788489 (rank : 21) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS18_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 32) | NC score | 0.761397 (rank : 34) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 33) | NC score | 0.762402 (rank : 32) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS12_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 34) | NC score | 0.737033 (rank : 38) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS12_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 35) | NC score | 0.731019 (rank : 40) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
DUS21_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 36) | NC score | 0.761599 (rank : 33) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 37) | NC score | 0.751406 (rank : 35) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 38) | NC score | 0.750756 (rank : 36) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS13_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 39) | NC score | 0.740569 (rank : 37) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS13_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 40) | NC score | 0.732832 (rank : 39) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
STYL1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 41) | NC score | 0.717371 (rank : 41) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 42) | NC score | 0.051882 (rank : 54) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 43) | NC score | 0.022730 (rank : 79) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PTPM1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 44) | NC score | 0.431434 (rank : 43) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.454689 (rank : 42) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.018777 (rank : 90) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.035392 (rank : 58) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.025238 (rank : 72) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.026796 (rank : 70) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.016876 (rank : 95) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.031031 (rank : 61) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.029690 (rank : 64) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.025495 (rank : 71) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
CC14A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.198889 (rank : 47) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.022583 (rank : 81) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SP7_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | -0.000487 (rank : 162) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VI67, Q8C5R3, Q8C6A7 | Gene names | Sp7, Osx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp7 (Zinc finger protein osterix) (C22). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.006989 (rank : 132) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.255854 (rank : 44) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
FA12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.001795 (rank : 158) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.027976 (rank : 67) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.027563 (rank : 69) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.188487 (rank : 49) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
EPM2A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.244313 (rank : 45) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.009309 (rank : 125) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
HN1L_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.029492 (rank : 65) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H910, Q6EIC7 | Gene names | C16orf34, HN1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1-like protein (HN1-like protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.015378 (rank : 101) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.012132 (rank : 110) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.014923 (rank : 104) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.021581 (rank : 84) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.038671 (rank : 57) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
ABCAD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.011094 (rank : 113) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
CM007_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.022067 (rank : 82) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.010904 (rank : 116) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
ZN358_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | -0.002602 (rank : 165) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
EPC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.013226 (rank : 107) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C9X6, Q9Z299 | Gene names | Epc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
PROM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.024752 (rank : 74) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54990, O35408 | Gene names | Prom1, Prom, Proml1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133 homolog). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.031814 (rank : 60) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.030582 (rank : 62) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.030352 (rank : 63) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.015337 (rank : 103) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.023642 (rank : 77) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PABP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.023739 (rank : 76) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86U42, O43484 | Gene names | PABPN1, PAB2, PABP2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
PABP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.023162 (rank : 78) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CCS6, O35935 | Gene names | Pabpn1, Pab2, Pabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.019497 (rank : 88) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.013724 (rank : 105) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.007065 (rank : 131) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.004033 (rank : 153) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.010184 (rank : 120) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
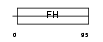
FOXL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.010971 (rank : 115) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.015786 (rank : 98) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.016044 (rank : 97) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.018065 (rank : 93) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.015655 (rank : 99) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TTK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | -0.002541 (rank : 164) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | -0.000991 (rank : 163) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.019827 (rank : 87) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.006868 (rank : 136) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | -0.002798 (rank : 166) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.024980 (rank : 73) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.005493 (rank : 142) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.018877 (rank : 89) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.016383 (rank : 96) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.002584 (rank : 156) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.005229 (rank : 143) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.009694 (rank : 123) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
HHIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.008650 (rank : 127) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
HHIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.009232 (rank : 126) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TN16, Q8C0B0, Q9WU59 | Gene names | Hhip, Hip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
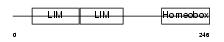
ISL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.002868 (rank : 155) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.033010 (rank : 59) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | -0.003153 (rank : 167) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.022720 (rank : 80) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.020865 (rank : 86) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.027688 (rank : 68) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
AP2A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.006664 (rank : 137) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.008589 (rank : 128) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.029030 (rank : 66) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CC14B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.210831 (rank : 46) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.007706 (rank : 130) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DUET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | -0.004199 (rank : 169) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.015547 (rank : 100) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.004251 (rank : 151) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
K1958_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.010211 (rank : 119) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
MKL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.010041 (rank : 121) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.012097 (rank : 111) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.015343 (rank : 102) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NELFA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.020994 (rank : 85) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.011167 (rank : 112) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.004680 (rank : 146) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
STAT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.005155 (rank : 144) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
STX12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.004512 (rank : 149) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.006890 (rank : 135) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.005949 (rank : 139) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
TP4A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.050604 (rank : 55) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.050604 (rank : 56) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.004618 (rank : 148) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.012294 (rank : 109) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
AMPH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.009755 (rank : 122) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
AP2A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.006962 (rank : 134) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.018313 (rank : 91) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.010708 (rank : 117) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.010366 (rank : 118) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.005778 (rank : 141) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.013344 (rank : 106) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
FOXK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.004747 (rank : 145) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
HTF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.005847 (rank : 140) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
MCL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.006989 (rank : 133) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97287, Q3TUS0, Q792P0, Q9CRI4 | Gene names | Mcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Bcl-2-related protein EAT/mcl1). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.012726 (rank : 108) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.004176 (rank : 152) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NARGL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.004671 (rank : 147) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.006309 (rank : 138) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.017940 (rank : 94) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.008063 (rank : 129) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.018124 (rank : 92) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NR4A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.001249 (rank : 160) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43354, Q16311 | Gene names | NR4A2, NOT, NURR1, TINUR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (Immediate-early response protein NOT) (Transcriptionally-inducible nuclear receptor). | |||||
|
NR4A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.001250 (rank : 159) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06219, O08690 | Gene names | Nr4a2, Nurr1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (NUR- related factor 1). | |||||
|
OSBL5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.003989 (rank : 154) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.000165 (rank : 161) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.021795 (rank : 83) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.024307 (rank : 75) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
STAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.011077 (rank : 114) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
TBCD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.004264 (rank : 150) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60343, Q5W0B9 | Gene names | TBC1D4, KIAA0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.009456 (rank : 124) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
VITRN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.002511 (rank : 157) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
ZXDB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | -0.003570 (rank : 168) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98169, Q9UBB3 | Gene names | ZXDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDB. | |||||
|
CC14B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.171463 (rank : 51) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.194649 (rank : 48) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
DUS23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.174217 (rank : 50) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
TP4A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.062104 (rank : 52) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.059323 (rank : 53) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 164 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.989644 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.965279 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.963383 (rank : 4) | θ value | 5.61992e-127 (rank : 5) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.962722 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.960700 (rank : 6) | θ value | 3.52827e-121 (rank : 6) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS19_HUMAN
|
||||||
| NC score | 0.830068 (rank : 7) | θ value | 6.62687e-19 (rank : 21) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS19_MOUSE
|
||||||
| NC score | 0.828316 (rank : 8) | θ value | 4.74913e-17 (rank : 26) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.813717 (rank : 9) | θ value | 3.75424e-22 (rank : 9) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.806299 (rank : 10) | θ value | 9.87957e-23 (rank : 7) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.806252 (rank : 11) | θ value | 9.87957e-23 (rank : 8) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
STYX_MOUSE
|
||||||
| NC score | 0.806080 (rank : 12) | θ value | 8.95645e-16 (rank : 29) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
DUS22_MOUSE
|
||||||
| NC score | 0.805437 (rank : 13) | θ value | 6.85773e-16 (rank : 28) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| NC score | 0.805401 (rank : 14) | θ value | 5.25075e-16 (rank : 27) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
STYX_HUMAN
|
||||||
| NC score | 0.804827 (rank : 15) | θ value | 4.44505e-15 (rank : 30) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.802236 (rank : 16) | θ value | 3.17815e-21 (rank : 13) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS14_MOUSE
|
||||||
| NC score | 0.799373 (rank : 17) | θ value | 1.13037e-18 (rank : 23) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS14_HUMAN
|
||||||
| NC score | 0.796504 (rank : 18) | θ value | 1.13037e-18 (rank : 22) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.795352 (rank : 19) | θ value | 1.2077e-20 (rank : 18) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.794348 (rank : 20) | θ value | 1.2077e-20 (rank : 17) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS15_HUMAN
|
||||||
| NC score | 0.788489 (rank : 21) | θ value | 7.09661e-13 (rank : 31) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS1_HUMAN
|
||||||
| NC score | 0.787749 (rank : 22) | θ value | 8.36355e-22 (rank : 10) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.787503 (rank : 23) | θ value | 9.24701e-21 (rank : 16) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.787240 (rank : 24) | θ value | 3.17815e-21 (rank : 14) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.785869 (rank : 25) | θ value | 1.2077e-20 (rank : 19) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS1_MOUSE
|
||||||
| NC score | 0.784427 (rank : 26) | θ value | 1.09232e-21 (rank : 11) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.781771 (rank : 27) | θ value | 1.42661e-21 (rank : 12) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.780882 (rank : 28) | θ value | 9.24701e-21 (rank : 15) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.775344 (rank : 29) | θ value | 5.60996e-18 (rank : 25) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.767827 (rank : 30) | θ value | 1.92812e-18 (rank : 24) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.762828 (rank : 31) | θ value | 5.07402e-19 (rank : 20) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.762402 (rank : 32) | θ value | 4.59992e-12 (rank : 33) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS21_HUMAN
|
||||||
| NC score | 0.761599 (rank : 33) | θ value | 2.98157e-11 (rank : 36) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_HUMAN
|
||||||
| NC score | 0.761397 (rank : 34) | θ value | 3.52202e-12 (rank : 32) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.751406 (rank : 35) | θ value | 9.59137e-10 (rank : 37) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| NC score | 0.750756 (rank : 36) | θ value | 1.63604e-09 (rank : 38) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS13_MOUSE
|
||||||
| NC score | 0.740569 (rank : 37) | θ value | 2.13673e-09 (rank : 39) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS12_MOUSE
|
||||||
| NC score | 0.737033 (rank : 38) | θ value | 1.33837e-11 (rank : 34) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS13_HUMAN
|
||||||
| NC score | 0.732832 (rank : 39) | θ value | 1.38499e-08 (rank : 40) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
DUS12_HUMAN
|
||||||
| NC score | 0.731019 (rank : 40) | θ value | 2.98157e-11 (rank : 35) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
STYL1_HUMAN
|
||||||
| NC score | 0.717371 (rank : 41) | θ value | 1.69304e-06 (rank : 41) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
PTPM1_MOUSE
|
||||||
| NC score | 0.454689 (rank : 42) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_HUMAN
|
||||||
| NC score | 0.431434 (rank : 43) | θ value | 0.0431538 (rank : 44) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.255854 (rank : 44) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_HUMAN
|
||||||
| NC score | 0.244313 (rank : 45) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.210831 (rank : 46) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14A_MOUSE
|
||||||
| NC score | 0.198889 (rank : 47) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
DUS23_HUMAN
|
||||||
| NC score | 0.194649 (rank : 48) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.188487 (rank : 49) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
DUS23_MOUSE
|
||||||
| NC score | 0.174217 (rank : 50) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
CC14B_MOUSE
|
||||||
| NC score | 0.171463 (rank : 51) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
TP4A3_HUMAN
|
||||||
| NC score | 0.062104 (rank : 52) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| NC score | 0.059323 (rank : 53) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.051882 (rank : 54) | θ value | 0.00869519 (rank : 42) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
TP4A1_HUMAN
|
||||||
| NC score | 0.050604 (rank : 55) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| NC score | 0.050604 (rank : 56) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.038671 (rank : 57) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.035392 (rank : 58) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.033010 (rank : 59) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.031814 (rank : 60) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.031031 (rank : 61) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.030582 (rank : 62) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.030352 (rank : 63) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.029690 (rank : 64) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
HN1L_HUMAN
|
||||||
| NC score | 0.029492 (rank : 65) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H910, Q6EIC7 | Gene names | C16orf34, HN1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1-like protein (HN1-like protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.029030 (rank : 66) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.027976 (rank : 67) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.027688 (rank : 68) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.027563 (rank : 69) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.026796 (rank : 70) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.025495 (rank : 71) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.025238 (rank : 72) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.024980 (rank : 73) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
PROM1_MOUSE
|
||||||
| NC score | 0.024752 (rank : 74) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54990, O35408 | Gene names | Prom1, Prom, Proml1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133 homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.024307 (rank : 75) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PABP2_HUMAN
|
||||||
| NC score | 0.023739 (rank : 76) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86U42, O43484 | Gene names | PABPN1, PAB2, PABP2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.023642 (rank : 77) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PABP2_MOUSE
|
||||||
| NC score | 0.023162 (rank : 78) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CCS6, O35935 | Gene names | Pabpn1, Pab2, Pabp2 | |||
|
Domain Architecture |

|
|||||
| Description | Polyadenylate-binding protein 2 (Poly(A)-binding protein 2) (Poly(A)- binding protein II) (PABII) (Polyadenylate-binding nuclear protein 1) (Nuclear poly(A)-binding protein 1). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.022730 (rank : 79) | θ value | 0.0252991 (rank : 43) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.022720 (rank : 80) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.022583 (rank : 81) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CM007_MOUSE
|
||||||
| NC score | 0.022067 (rank : 82) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K2Y0, Q9CYU1, Q9D1Y0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf7 homolog. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.021795 (rank : 83) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.021581 (rank : 84) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NELFA_HUMAN
|
||||||
| NC score | 0.020994 (rank : 85) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3P2, O95392 | Gene names | WHSC2, NELFA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.020865 (rank : 86) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.019827 (rank : 87) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.019497 (rank : 88) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.018877 (rank : 89) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.018777 (rank : 90) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.018313 (rank : 91) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.018124 (rank : 92) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.018065 (rank : 93) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.017940 (rank : 94) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.016876 (rank : 95) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.016383 (rank : 96) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.016044 (rank : 97) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.015786 (rank : 98) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.015655 (rank : 99) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.015547 (rank : 100) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.015378 (rank : 101) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.015343 (rank : 102) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.015337 (rank : 103) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.014923 (rank : 104) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.013724 (rank : 105) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.013344 (rank : 106) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
EPC1_MOUSE
|
||||||
| NC score | 0.013226 (rank : 107) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C9X6, Q9Z299 | Gene names | Epc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.012726 (rank : 108) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.012294 (rank : 109) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.012132 (rank : 110) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.012097 (rank : 111) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.011167 (rank : 112) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
ABCAD_HUMAN
|
||||||
| NC score | 0.011094 (rank : 113) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UQ4, Q6ZTT7, Q86WI2, Q8N248 | Gene names | ABCA13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
STAP2_MOUSE
|
||||||
| NC score | 0.011077 (rank : 114) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R0L1, Q8BWS2 | Gene names | Stap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-transducing adaptor protein 2 (STAP-2). | |||||
|
FOXL1_MOUSE
|
||||||
| NC score | 0.010971 (rank : 115) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.010904 (rank : 116) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.010708 (rank : 117) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| NC score | 0.010366 (rank : 118) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
K1958_HUMAN
|
||||||
| NC score | 0.010211 (rank : 119) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8K9, Q5T252, Q8TF43, Q96N02 | Gene names | KIAA1958 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.010184 (rank : 120) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
MKL2_HUMAN
|
||||||
| NC score | 0.010041 (rank : 121) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
AMPH_HUMAN
|
||||||
| NC score | 0.009755 (rank : 122) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49418, O43538, Q75MJ8, Q75MK5, Q75MM3, Q8N4G0 | Gene names | AMPH, AMPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Amphiphysin. | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.009694 (rank : 123) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.009456 (rank : 124) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.009309 (rank : 125) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
HHIP_MOUSE
|
||||||
| NC score | 0.009232 (rank : 126) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TN16, Q8C0B0, Q9WU59 | Gene names | Hhip, Hip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
HHIP_HUMAN
|
||||||
| NC score | 0.008650 (rank : 127) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.008589 (rank : 128) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.008063 (rank : 129) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.007706 (rank : 130) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.007065 (rank : 131) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.006989 (rank : 132) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
MCL1_MOUSE
|
||||||
| NC score | 0.006989 (rank : 133) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97287, Q3TUS0, Q792P0, Q9CRI4 | Gene names | Mcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Bcl-2-related protein EAT/mcl1). | |||||
|
AP2A1_HUMAN
|
||||||
| NC score | 0.006962 (rank : 134) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95782, Q96CI7, Q96PP6, Q96PP7, Q9H070 | Gene names | AP2A1, ADTAA | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.006890 (rank : 135) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.006868 (rank : 136) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
AP2A1_MOUSE
|
||||||
| NC score | 0.006664 (rank : 137) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17426 | Gene names | Ap2a1, Adtaa, Clapa1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-1 (Adapter-related protein complex 2 alpha- 1 subunit) (Alpha-adaptin A) (Adaptor protein complex AP-2 alpha-1 subunit) (Clathrin assembly protein complex 2 alpha-A large chain) (100 kDa coated vesicle protein A) (Plasma membrane adaptor HA2/AP2 adaptin alpha A subunit). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.006309 (rank : 138) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.005949 (rank : 139) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
HTF4_MOUSE
|
||||||
| NC score | 0.005847 (rank : 140) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61286 | Gene names | Tcf12, Alf1, Meb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4) (Class A helix-loop-helix transcription factor ME1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.005778 (rank : 141) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.005493 (rank : 142) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.005229 (rank : 143) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
STAT2_MOUSE
|
||||||
| NC score | 0.005155 (rank : 144) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVL2, Q64188, Q64189, Q64250 | Gene names | Stat2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 2. | |||||
|
FOXK2_HUMAN
|
||||||
| NC score | 0.004747 (rank : 145) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01167, Q13622, Q13623, Q13624 | Gene names | FOXK2, ILF, ILF1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein K2 (Interleukin enhancer-binding factor 1) (Cellular transcription factor ILF-1). | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.004680 (rank : 146) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
NARGL_MOUSE
|
||||||
| NC score | 0.004671 (rank : 147) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBB4, Q3U7V2 | Gene names | Narg1l, Nat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NMDA receptor-regulated 1-like protein (NARG1-like protein). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.004618 (rank : 148) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
STX12_HUMAN
|
||||||
| NC score | 0.004512 (rank : 149) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
TBCD4_HUMAN
|
||||||
| NC score | 0.004264 (rank : 150) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60343, Q5W0B9 | Gene names | TBC1D4, KIAA0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.004251 (rank : 151) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.004176 (rank : 152) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.004033 (rank : 153) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
OSBL5_MOUSE
|
||||||
| NC score | 0.003989 (rank : 154) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER64, Q8R510, Q99NF5 | Gene names | Osbpl5, Obph1, Osbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 5 (OSBP-related protein 5) (ORP-5) (Oxysterol-binding protein homolog 1). | |||||
|
ISL2_HUMAN
|
||||||
| NC score | 0.002868 (rank : 155) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.002584 (rank : 156) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
VITRN_MOUSE
|
||||||
| NC score | 0.002511 (rank : 157) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.001795 (rank : 158) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
NR4A2_MOUSE
|
||||||
| NC score | 0.001250 (rank : 159) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06219, O08690 | Gene names | Nr4a2, Nurr1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (NUR- related factor 1). | |||||
|
NR4A2_HUMAN
|
||||||
| NC score | 0.001249 (rank : 160) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43354, Q16311 | Gene names | NR4A2, NOT, NURR1, TINUR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (Immediate-early response protein NOT) (Transcriptionally-inducible nuclear receptor). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.000165 (rank : 161) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SP7_MOUSE
|
||||||
| NC score | -0.000487 (rank : 162) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 756 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VI67, Q8C5R3, Q8C6A7 | Gene names | Sp7, Osx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp7 (Zinc finger protein osterix) (C22). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | -0.000991 (rank : 163) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
TTK_MOUSE
|
||||||
| NC score | -0.002541 (rank : 164) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35761 | Gene names | Ttk, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein kinase TTK (EC 2.7.12.1) (ESK) (PYT). | |||||
|
ZN358_HUMAN
|
||||||
| NC score | -0.002602 (rank : 165) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | -0.002798 (rank : 166) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | -0.003153 (rank : 167) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
ZXDB_HUMAN
|
||||||
| NC score | -0.003570 (rank : 168) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98169, Q9UBB3 | Gene names | ZXDB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger X-linked protein ZXDB. | |||||
|
DUET_HUMAN
|
||||||
| NC score | -0.004199 (rank : 169) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 164 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||