Please be patient as the page loads
|
DUS8_MOUSE
|
||||||
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DUS8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984860 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 5.80223e-132 (rank : 3) | NC score | 0.975594 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS10_HUMAN
|
||||||
| θ value | 3.72821e-46 (rank : 4) | NC score | 0.930432 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_MOUSE
|
||||||
| θ value | 2.41655e-45 (rank : 5) | NC score | 0.931292 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS7_HUMAN
|
||||||
| θ value | 9.50281e-42 (rank : 6) | NC score | 0.926026 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| θ value | 3.99248e-40 (rank : 7) | NC score | 0.922434 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS1_MOUSE
|
||||||
| θ value | 8.89434e-40 (rank : 8) | NC score | 0.914573 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS6_MOUSE
|
||||||
| θ value | 1.16164e-39 (rank : 9) | NC score | 0.922032 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS4_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 10) | NC score | 0.913212 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS9_HUMAN
|
||||||
| θ value | 3.37985e-39 (rank : 11) | NC score | 0.932918 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS4_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 12) | NC score | 0.913939 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS1_HUMAN
|
||||||
| θ value | 2.86122e-38 (rank : 13) | NC score | 0.914311 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS2_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 14) | NC score | 0.921718 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 15) | NC score | 0.921269 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS5_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 16) | NC score | 0.911789 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
SSH3_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 17) | NC score | 0.786757 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH1_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 18) | NC score | 0.767827 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 19) | NC score | 0.782892 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
STYX_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 20) | NC score | 0.860156 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
STYX_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 21) | NC score | 0.861252 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 22) | NC score | 0.751755 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
SSH2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 23) | NC score | 0.753891 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
DUS22_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 24) | NC score | 0.865897 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS22_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 25) | NC score | 0.866232 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
SSH1_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 26) | NC score | 0.767494 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
DUS14_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 27) | NC score | 0.855894 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS18_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 28) | NC score | 0.837548 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS18_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 29) | NC score | 0.837538 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS14_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 30) | NC score | 0.857234 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS19_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 31) | NC score | 0.862377 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS21_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 32) | NC score | 0.838108 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS19_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 33) | NC score | 0.856812 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS15_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 34) | NC score | 0.846555 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS12_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 35) | NC score | 0.782819 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS3_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 36) | NC score | 0.825534 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 37) | NC score | 0.824325 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
DUS12_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 38) | NC score | 0.766542 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
DUS13_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 39) | NC score | 0.782199 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS13_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 40) | NC score | 0.777005 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 41) | NC score | 0.065910 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
STYL1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 42) | NC score | 0.772812 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 43) | NC score | 0.039655 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
DUS23_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 44) | NC score | 0.341671 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
DUS23_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 45) | NC score | 0.315096 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
PTPM1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 46) | NC score | 0.516614 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
CC14B_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 47) | NC score | 0.319878 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 48) | NC score | 0.027720 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.026239 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PTPM1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.479907 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 51) | NC score | 0.045590 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 52) | NC score | 0.026693 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 53) | NC score | 0.049179 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
DUS11_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 54) | NC score | 0.112388 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
CC14B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 55) | NC score | 0.281833 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 56) | NC score | 0.028472 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
TP4A2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.130017 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12974, O00649, Q15197, Q15259, Q15260, Q15261 | Gene names | PTP4A2, PRL2, PTPCAAX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2) (PTP(CAAXII)) (HU-PP-1) (OV-1). | |||||
|
TP4A2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 58) | NC score | 0.130017 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70274, Q3U1K7 | Gene names | Ptp4a2, Prl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2). | |||||
|
EPM2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 59) | NC score | 0.282969 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | 0.284199 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.014033 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.026101 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.020088 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
TP4A1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 64) | NC score | 0.131268 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.131268 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.027381 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.021802 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SENP5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.023844 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.001413 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
TP4A3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.158243 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 71) | NC score | 0.154280 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
FGD3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.011326 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.018279 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 74) | NC score | 0.035019 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CC14A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.257377 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.260824 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.019436 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.034605 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.033028 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.035784 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.008658 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.020969 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.031442 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.026460 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 85) | NC score | 0.040012 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.001898 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ICAM4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.027399 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14773, Q14771, Q14772, Q16375 | Gene names | ICAM4, LW | |||
|
Domain Architecture |
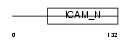
|
|||||
| Description | Intercellular adhesion molecule 4 precursor (ICAM-4) (Landsteiner- Wiener blood group glycoprotein) (LW blood group protein) (CD242 antigen). | |||||
|
P73_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.013734 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.012484 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.017097 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.017636 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.017277 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
NR2E3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.002432 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |
|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.018296 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.010882 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.018836 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.009458 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.015296 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
GDF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.004418 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20863 | Gene names | Gdf1, Gdf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Embryonic growth/differentiation factor 1 precursor (GDF-1). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.006002 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.010537 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
BC11B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | -0.004055 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0K0, Q9H162 | Gene names | BCL11B, CTIP2, RIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (hRit1) (COUP-TF-interacting protein 2). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.013648 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CD248_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.005258 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
ELL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.012431 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.007166 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PTPRE_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.017476 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.001312 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.021807 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.014894 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.007702 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
PTPRE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.013777 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
CB024_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.008096 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K158, Q3TE87 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf24 homolog. | |||||
|
CDX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | -0.001330 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14627, Q5JS20 | Gene names | CDX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.004340 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CF015_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.015642 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
NOL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.015048 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
|
|||||
| Description | Nucleolar protein 3. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.008309 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PTPRA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.015744 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
DUS11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.054183 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75319, Q6AI47, Q9BWE3 | Gene names | DUSP11, PIR1 | |||
|
Domain Architecture |
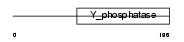
|
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.055447 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.051168 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.056999 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.059359 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.984860 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.975594 (rank : 3) | θ value | 5.80223e-132 (rank : 3) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
DUS9_HUMAN
|
||||||
| NC score | 0.932918 (rank : 4) | θ value | 3.37985e-39 (rank : 11) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99956 | Gene names | DUSP9, MKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 9 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 4) (MAP kinase phosphatase 4) (MKP-4). | |||||
|
DUS10_MOUSE
|
||||||
| NC score | 0.931292 (rank : 5) | θ value | 2.41655e-45 (rank : 5) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESS0, Q9CZY9 | Gene names | Dusp10, Mkp5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS10_HUMAN
|
||||||
| NC score | 0.930432 (rank : 6) | θ value | 3.72821e-46 (rank : 4) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Y6W6 | Gene names | DUSP10, MKP5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 10 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 5) (MAP kinase phosphatase 5) (MKP-5). | |||||
|
DUS7_HUMAN
|
||||||
| NC score | 0.926026 (rank : 7) | θ value | 9.50281e-42 (rank : 6) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q16829, Q8NFJ0 | Gene names | DUSP7, PYST2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 7 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PYST2). | |||||
|
DUS6_HUMAN
|
||||||
| NC score | 0.922434 (rank : 8) | θ value | 3.99248e-40 (rank : 7) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16828, O75109, Q53Y75, Q9BSH6 | Gene names | DUSP6, MKP3, PYST1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3) (Dual specificity protein phosphatase PYST1). | |||||
|
DUS6_MOUSE
|
||||||
| NC score | 0.922032 (rank : 9) | θ value | 1.16164e-39 (rank : 9) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBB1, Q542I5, Q9D7L4 | Gene names | Dusp6, Mkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 6 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 3) (MAP kinase phosphatase 3) (MKP-3). | |||||
|
DUS2_MOUSE
|
||||||
| NC score | 0.921718 (rank : 10) | θ value | 4.568e-36 (rank : 14) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q05922, Q60640 | Gene names | Dusp2, Pac-1, Pac1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS2_HUMAN
|
||||||
| NC score | 0.921269 (rank : 11) | θ value | 5.96599e-36 (rank : 15) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q05923 | Gene names | DUSP2, PAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 2 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase PAC-1). | |||||
|
DUS1_MOUSE
|
||||||
| NC score | 0.914573 (rank : 12) | θ value | 8.89434e-40 (rank : 8) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28563 | Gene names | Dusp1, 3ch134, Mkp1, Ptpn10, Ptpn16 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase 3CH134) (Protein-tyrosine phosphatase ERP). | |||||
|
DUS1_HUMAN
|
||||||
| NC score | 0.914311 (rank : 13) | θ value | 2.86122e-38 (rank : 13) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P28562, Q2V508 | Gene names | DUSP1, CL100, MKP1, PTPN10, VH1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 1 (EC 3.1.3.48) (EC 3.1.3.16) (MAP kinase phosphatase 1) (MKP-1) (Protein-tyrosine phosphatase CL100) (Dual specificity protein phosphatase hVH1). | |||||
|
DUS4_HUMAN
|
||||||
| NC score | 0.913939 (rank : 14) | θ value | 4.41421e-39 (rank : 12) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q13115, Q13524 | Gene names | DUSP4, MKP2, VH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 2) (MAP kinase phosphatase 2) (MKP-2) (Dual specificity protein phosphatase hVH2). | |||||
|
DUS4_MOUSE
|
||||||
| NC score | 0.913212 (rank : 15) | θ value | 3.37985e-39 (rank : 10) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BFV3 | Gene names | Dusp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 4 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS5_HUMAN
|
||||||
| NC score | 0.911789 (rank : 16) | θ value | 1.28732e-30 (rank : 16) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q16690, Q12997 | Gene names | DUSP5, VH3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 5 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH3). | |||||
|
DUS22_MOUSE
|
||||||
| NC score | 0.866232 (rank : 17) | θ value | 3.63628e-17 (rank : 25) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99N11, Q5SQN9, Q5SQP0 | Gene names | Dusp22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (LMW-DSP2). | |||||
|
DUS22_HUMAN
|
||||||
| NC score | 0.865897 (rank : 18) | θ value | 2.13179e-17 (rank : 24) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NRW4, Q59GW2, Q5VWR2, Q96AR1 | Gene names | DUSP22, JSP1, MKPX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 22 (EC 3.1.3.48) (EC 3.1.3.16) (JNK-stimulatory phosphatase-1) (JSP-1) (Mitogen-activated protein kinase phosphatase x) (LMW-DSP2). | |||||
|
DUS19_HUMAN
|
||||||
| NC score | 0.862377 (rank : 19) | θ value | 1.99529e-15 (rank : 31) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WTR2, Q8WYN4 | Gene names | DUSP19, DUSP17, SKRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
STYX_HUMAN
|
||||||
| NC score | 0.861252 (rank : 20) | θ value | 5.60996e-18 (rank : 21) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8WUJ0, Q99850 | Gene names | STYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein). | |||||
|
STYX_MOUSE
|
||||||
| NC score | 0.860156 (rank : 21) | θ value | 4.29542e-18 (rank : 20) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q60969, Q3TMA3, Q60970, Q9DCF8 | Gene names | Styx | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting protein (Protein tyrosine phosphatase-like protein) (Phosphoserine/threonine/tyrosine interaction protein). | |||||
|
DUS14_MOUSE
|
||||||
| NC score | 0.857234 (rank : 22) | θ value | 5.25075e-16 (rank : 30) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JLY7, Q9D715 | Gene names | Dusp14, Mkp6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6). | |||||
|
DUS19_MOUSE
|
||||||
| NC score | 0.856812 (rank : 23) | θ value | 5.8054e-15 (rank : 33) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8K4T5 | Gene names | Dusp19, Skrp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 19 (EC 3.1.3.48) (EC 3.1.3.16) (Stress-activated protein kinase pathway-regulating phosphatase 1) (Protein phosphatase SKRP1). | |||||
|
DUS14_HUMAN
|
||||||
| NC score | 0.855894 (rank : 24) | θ value | 1.38178e-16 (rank : 27) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95147 | Gene names | DUSP14, MKP6 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 14 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 6) (MAP kinase phosphatase 6) (MKP-6) (MKP-1-like protein tyrosine phosphatase) (MKP- L). | |||||
|
DUS15_HUMAN
|
||||||
| NC score | 0.846555 (rank : 25) | θ value | 1.68911e-14 (rank : 34) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H1R2, Q5QP62, Q5QP63, Q5QP65, Q6PGN7, Q8N826, Q9BX24 | Gene names | DUSP15, C20orf57, VHY | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 15 (EC 3.1.3.48) (EC 3.1.3.16) (Vaccinia virus VH1-related dual-specific protein phosphatase Y) (VH1- related member Y). | |||||
|
DUS21_HUMAN
|
||||||
| NC score | 0.838108 (rank : 26) | θ value | 1.99529e-15 (rank : 32) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H596, Q6IAJ6, Q6YDQ8 | Gene names | DUSP21, LMWDSP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 21 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 21). | |||||
|
DUS18_MOUSE
|
||||||
| NC score | 0.837548 (rank : 27) | θ value | 2.35696e-16 (rank : 28) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8VE01 | Gene names | Dusp18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16). | |||||
|
DUS18_HUMAN
|
||||||
| NC score | 0.837538 (rank : 28) | θ value | 3.07829e-16 (rank : 29) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8NEJ0 | Gene names | DUSP18, LMWDSP20 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 18 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular weight dual specificity phosphatase 20). | |||||
|
DUS3_HUMAN
|
||||||
| NC score | 0.825534 (rank : 29) | θ value | 1.133e-10 (rank : 36) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P51452 | Gene names | DUSP3, VHR | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase VHR). | |||||
|
DUS3_MOUSE
|
||||||
| NC score | 0.824325 (rank : 30) | θ value | 2.52405e-10 (rank : 37) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D7X3 | Gene names | Dusp3 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 3 (EC 3.1.3.48) (EC 3.1.3.16) (T- DSP11). | |||||
|
SSH3_MOUSE
|
||||||
| NC score | 0.786757 (rank : 31) | θ value | 2.06002e-20 (rank : 17) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K330, Q3UDX0 | Gene names | Ssh3, Ssh3l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (mSSH-3L). | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.782892 (rank : 32) | θ value | 2.5182e-18 (rank : 19) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DUS12_MOUSE
|
||||||
| NC score | 0.782819 (rank : 33) | θ value | 1.74796e-11 (rank : 35) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9D0T2, Q9EQD3 | Gene names | Dusp12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity phosphatase T-DSP4) (Dual specificity phosphatase VH1). | |||||
|
DUS13_MOUSE
|
||||||
| NC score | 0.782199 (rank : 34) | θ value | 6.21693e-09 (rank : 39) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QYJ7 | Gene names | Dusp13, Tmdp | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal muscle-specific DSP) (Dual specificity tyrosine phosphatase TS-DSP6). | |||||
|
DUS13_HUMAN
|
||||||
| NC score | 0.777005 (rank : 35) | θ value | 4.0297e-08 (rank : 40) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UII6, Q5JSC6, Q96GC2 | Gene names | DUSP13, TMDP | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 13 (EC 3.1.3.48) (EC 3.1.3.16) (Testis- and skeletal-muscle-specific DSP) (Dual specificity phosphatase SKRP4). | |||||
|
STYL1_HUMAN
|
||||||
| NC score | 0.772812 (rank : 36) | θ value | 2.88788e-06 (rank : 42) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6J8, Q9UBP1, Q9UK06, Q9UK07, Q9UKG2, Q9UKG3 | Gene names | STYXL1, DUSP24, MKSTYX | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine/tyrosine-interacting-like protein 1 (Dual specificity protein phosphatase 24) (Map kinase phosphatase-like protein MK-STYX) (Dual specificity phosphatase inhibitor MK-STYX). | |||||
|
SSH1_HUMAN
|
||||||
| NC score | 0.767827 (rank : 37) | θ value | 1.92812e-18 (rank : 18) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WYL5, Q6P6C0, Q8N9A7, Q8WYL3, Q8WYL4, Q9P2P8 | Gene names | SSH1, KIAA1298, SSH1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (hSSH-1L). | |||||
|
SSH1_MOUSE
|
||||||
| NC score | 0.767494 (rank : 38) | θ value | 4.74913e-17 (rank : 26) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q76I79, Q3TDG3, Q69ZM4, Q811E5 | Gene names | Ssh1, Kiaa1298, Ssh1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 1 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-1L) (mSSH-1L). | |||||
|
DUS12_HUMAN
|
||||||
| NC score | 0.766542 (rank : 39) | θ value | 5.62301e-10 (rank : 38) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UNI6 | Gene names | DUSP12 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 12 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity tyrosine phosphatase YVH1). | |||||
|
SSH2_MOUSE
|
||||||
| NC score | 0.753891 (rank : 40) | θ value | 1.24977e-17 (rank : 23) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SW75, Q3TDK8, Q3TYP8, Q3U2K3, Q5F268, Q5SW74, Q69ZC3, Q76I78 | Gene names | Ssh2, Kiaa1725, Ssh2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (mSSH-2L). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.751755 (rank : 41) | θ value | 1.24977e-17 (rank : 22) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
PTPM1_MOUSE
|
||||||
| NC score | 0.516614 (rank : 42) | θ value | 0.00134147 (rank : 46) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q66GT5, Q9CSJ8, Q9D622 | Gene names | Ptpmt1, Plip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
PTPM1_HUMAN
|
||||||
| NC score | 0.479907 (rank : 43) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8WUK0, Q7Z557, Q96CR2, Q9BXV8 | Gene names | PTPMT1, MOSP, PLIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor (EC 3.1.3.48) (EC 3.1.3.16) (PTEN-like phosphatase) (Phosphoinositide lipid phosphatase). | |||||
|
DUS23_HUMAN
|
||||||
| NC score | 0.341671 (rank : 44) | θ value | 0.000270298 (rank : 44) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BVJ7, Q9NX48 | Gene names | DUSP23, LDP3, VHZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3) (VH1-like phosphatase Z). | |||||
|
CC14B_HUMAN
|
||||||
| NC score | 0.319878 (rank : 45) | θ value | 0.00390308 (rank : 47) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O60729, O43183, O60730 | Gene names | CDC14B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
DUS23_MOUSE
|
||||||
| NC score | 0.315096 (rank : 46) | θ value | 0.00102713 (rank : 45) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6NT99, Q9CW48 | Gene names | Dusp23, Ldp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase 23 (EC 3.1.3.48) (EC 3.1.3.16) (Low molecular mass dual specificity phosphatase 3) (LDP-3). | |||||
|
EPM2A_MOUSE
|
||||||
| NC score | 0.284199 (rank : 47) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WUA5, Q8BY80 | Gene names | Epm2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
EPM2A_HUMAN
|
||||||
| NC score | 0.282969 (rank : 48) | θ value | 0.163984 (rank : 59) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95278, O95483, Q8IU96, Q8IX24, Q8IX25, Q9BS66, Q9UEN2 | Gene names | EPM2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Laforin (EC 3.1.3.48) (EC 3.1.3.16) (Lafora PTPase) (LAFPTPase). | |||||
|
CC14B_MOUSE
|
||||||
| NC score | 0.281833 (rank : 49) | θ value | 0.0563607 (rank : 55) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6PFY9, Q80WC4, Q8BLV5 | Gene names | Cdc14b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14B (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog B). | |||||
|
CC14A_MOUSE
|
||||||
| NC score | 0.260824 (rank : 50) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6GQT0, Q8BZ66 | Gene names | Cdc14a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
CC14A_HUMAN
|
||||||
| NC score | 0.257377 (rank : 51) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UNH5, O43171, O60727, O60728, Q8IXX0 | Gene names | CDC14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity protein phosphatase CDC14A (EC 3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14 homolog A). | |||||
|
TP4A3_HUMAN
|
||||||
| NC score | 0.158243 (rank : 52) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75365, Q8IVN5, Q99849, Q9BTW5 | Gene names | PTP4A3, PRL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3) (PRL-R). | |||||
|
TP4A3_MOUSE
|
||||||
| NC score | 0.154280 (rank : 53) | θ value | 0.365318 (rank : 71) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9D658, O70275, Q3T9Z5, Q9CTC8 | Gene names | Ptp4a3, Prl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 3 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a3) (Protein-tyrosine phosphatase of regenerating liver 3) (PRL-3). | |||||
|
TP4A1_HUMAN
|
||||||
| NC score | 0.131268 (rank : 54) | θ value | 0.21417 (rank : 64) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q93096, O00648, Q49A54 | Gene names | PTP4A1, PRL1, PTPCAAX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1) (PTP(CAAXI)). | |||||
|
TP4A1_MOUSE
|
||||||
| NC score | 0.131268 (rank : 55) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q63739, O09097, O09154, Q3UFU9 | Gene names | Ptp4a1, Prl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a1) (Protein-tyrosine phosphatase of regenerating liver 1) (PRL-1). | |||||
|
TP4A2_HUMAN
|
||||||
| NC score | 0.130017 (rank : 56) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12974, O00649, Q15197, Q15259, Q15260, Q15261 | Gene names | PTP4A2, PRL2, PTPCAAX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2) (PTP(CAAXII)) (HU-PP-1) (OV-1). | |||||
|
TP4A2_MOUSE
|
||||||
| NC score | 0.130017 (rank : 57) | θ value | 0.0961366 (rank : 58) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70274, Q3U1K7 | Gene names | Ptp4a2, Prl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine phosphatase type IVA protein 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase 4a2) (Protein-tyrosine phosphatase of regenerating liver 2) (PRL-2). | |||||
|
DUS11_MOUSE
|
||||||
| NC score | 0.112388 (rank : 58) | θ value | 0.0330416 (rank : 54) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NXK5, Q8BTR4, Q8BYE4 | Gene names | Dusp11, Pir1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.065910 (rank : 59) | θ value | 1.29631e-06 (rank : 41) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.059359 (rank : 60) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.056999 (rank : 61) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.055447 (rank : 62) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
DUS11_HUMAN
|
||||||
| NC score | 0.054183 (rank : 63) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75319, Q6AI47, Q9BWE3 | Gene names | DUSP11, PIR1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA/RNP complex-1-interacting phosphatase (EC 3.1.3.-) (Phosphatase that interacts with RNA/RNP complex 1) (Dual specificity protein phosphatase 11). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.051168 (rank : 64) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.049179 (rank : 65) | θ value | 0.0252991 (rank : 53) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.045590 (rank : 66) | θ value | 0.0193708 (rank : 51) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.040012 (rank : 67) | θ value | 1.38821 (rank : 85) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.039655 (rank : 68) | θ value | 0.00020696 (rank : 43) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.035784 (rank : 69) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.035019 (rank : 70) | θ value | 0.62314 (rank : 74) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.034605 (rank : 71) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.033028 (rank : 72) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.031442 (rank : 73) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.028472 (rank : 74) | θ value | 0.0736092 (rank : 56) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.027720 (rank : 75) | θ value | 0.00665767 (rank : 48) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ICAM4_HUMAN
|
||||||
| NC score | 0.027399 (rank : 76) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14773, Q14771, Q14772, Q16375 | Gene names | ICAM4, LW | |||
|
Domain Architecture |

|
|||||
| Description | Intercellular adhesion molecule 4 precursor (ICAM-4) (Landsteiner- Wiener blood group glycoprotein) (LW blood group protein) (CD242 antigen). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.027381 (rank : 77) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.026693 (rank : 78) | θ value | 0.0252991 (rank : 52) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.026460 (rank : 79) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.026239 (rank : 80) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.026101 (rank : 81) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SENP5_HUMAN
|
||||||
| NC score | 0.023844 (rank : 82) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.021807 (rank : 83) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.021802 (rank : 84) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.020969 (rank : 85) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.020088 (rank : 86) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.019436 (rank : 87) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.018836 (rank : 88) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.018296 (rank : 89) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.018279 (rank : 90) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.017636 (rank : 91) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
PTPRE_HUMAN
|
||||||
| NC score | 0.017476 (rank : 92) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23469, Q96KQ6 | Gene names | PTPRE | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.017277 (rank : 93) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.017097 (rank : 94) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
PTPRA_MOUSE
|
||||||
| NC score | 0.015744 (rank : 95) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18052, Q61808 | Gene names | Ptpra, Lrp, Ptpa | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase alpha precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase alpha) (R-PTP-alpha) (LCA- related phosphatase) (PTPTY-28). | |||||
|
CF015_HUMAN
|
||||||
| NC score | 0.015642 (rank : 96) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.015296 (rank : 97) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.015048 (rank : 98) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.014894 (rank : 99) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.014033 (rank : 100) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
PTPRE_MOUSE
|
||||||
| NC score | 0.013777 (rank : 101) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49446, Q62134, Q62444, Q64496 | Gene names | Ptpre, Ptpe | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase epsilon precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase epsilon) (R-PTP-epsilon). | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.013734 (rank : 102) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.013648 (rank : 103) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.012484 (rank : 104) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
ELL_MOUSE
|
||||||
| NC score | 0.012431 (rank : 105) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
FGD3_MOUSE
|
||||||
| NC score | 0.011326 (rank : 106) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88842, Q8BQ72 | Gene names | Fgd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 3. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.010882 (rank : 107) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.010537 (rank : 108) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.009458 (rank : 109) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.008658 (rank : 110) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.008309 (rank : 111) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
CB024_MOUSE
|
||||||
| NC score | 0.008096 (rank : 112) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K158, Q3TE87 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf24 homolog. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.007702 (rank : 113) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.007166 (rank : 114) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.006002 (rank : 115) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.005258 (rank : 116) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
GDF1_MOUSE
|
||||||
| NC score | 0.004418 (rank : 117) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20863 | Gene names | Gdf1, Gdf-1 | |||
|
Domain Architecture |

|
|||||
| Description | Embryonic growth/differentiation factor 1 precursor (GDF-1). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.004340 (rank : 118) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
NR2E3_MOUSE
|
||||||
| NC score | 0.002432 (rank : 119) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |

|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.001898 (rank : 120) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.001413 (rank : 121) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.001312 (rank : 122) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CDX4_HUMAN
|
||||||
| NC score | -0.001330 (rank : 123) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14627, Q5JS20 | Gene names | CDX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
BC11B_HUMAN
|
||||||
| NC score | -0.004055 (rank : 124) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 119 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0K0, Q9H162 | Gene names | BCL11B, CTIP2, RIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (hRit1) (COUP-TF-interacting protein 2). | |||||