Please be patient as the page loads
|
SENP5_HUMAN
|
||||||
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SENP5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
SENP3_MOUSE
|
||||||
| θ value | 3.81361e-83 (rank : 2) | NC score | 0.926753 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EP97 | Gene names | Senp3, Smt3ip, Smt3ip1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1). | |||||
|
SENP3_HUMAN
|
||||||
| θ value | 4.98072e-83 (rank : 3) | NC score | 0.928583 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
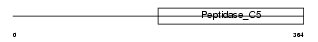
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |
|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
SENP1_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 4) | NC score | 0.837669 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP1_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 5) | NC score | 0.832687 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP2_HUMAN
|
||||||
| θ value | 5.584e-34 (rank : 6) | NC score | 0.812454 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HC62, Q96SR2, Q9P2L5 | Gene names | SENP2, KIAA1331 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SMT3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP2_MOUSE
|
||||||
| θ value | 2.77131e-33 (rank : 7) | NC score | 0.812624 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZX6, Q544T8, Q9D4Z0 | Gene names | Senp2, Smt3ip2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP7_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 8) | NC score | 0.473921 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
|
SENP6_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 9) | NC score | 0.489656 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZR1, O94891, Q8TBY4, Q9UJV5 | Gene names | SENP6, KIAA0797, SSP1, SUSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 6 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP6) (SUMO-1-specific protease 1). | |||||
|
SENP8_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 10) | NC score | 0.296252 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96LD8, Q96QA4 | Gene names | SENP8, DEN1, NEDP1, PRSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (Protease, cysteine 2) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
SENP8_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 11) | NC score | 0.260246 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
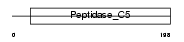
| SwissProt Accessions | Q9D2Z4, Q543P0 | Gene names | Senp8, Den1, Nedp1 | |||
|
Domain Architecture |
|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
RBNS5_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.048953 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.029915 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.023844 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.031952 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
LRRC7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.016545 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.025933 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.026100 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.015565 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PTHR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.013317 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41593, Q62119 | Gene names | Pthr1, Pthr | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor (PTH/PTHr receptor) (PTH/PTHrP type I receptor). | |||||
|
LRIG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.005879 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JA1, Q6IQ51, Q96CF9, Q9BYB8, Q9UFI4 | Gene names | LRIG1, LIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.013750 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.005171 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.016386 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.015228 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.018649 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CENPK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.022812 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESN5, Q3UXA9, Q569Q3, Q8C469 | Gene names | Cenpk, Solt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (SoxLZ/Sox6 leucine zipper-binding protein in testis). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.022440 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
CSRP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.012642 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.012569 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
K1B24_MOUSE
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.002542 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
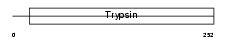
| SwissProt Accessions | Q61754, Q61755, Q9JM69 | Gene names | Klk1b24, Klk-24, Klk24 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b24 precursor (EC 3.4.21.35) (Glandular kallikrein K24) (Tissue kallikrein 24) (mGK-24). | |||||
|
CABYR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.017974 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
IFI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.008932 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
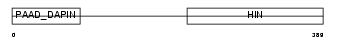
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.012632 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.012640 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.008843 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TM171_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.015191 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVE6, Q8N0S1, Q8TDT7 | Gene names | TMEM171 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 171. | |||||
|
SENP5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
SENP3_HUMAN
|
||||||
| NC score | 0.928583 (rank : 2) | θ value | 4.98072e-83 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
SENP3_MOUSE
|
||||||
| NC score | 0.926753 (rank : 3) | θ value | 3.81361e-83 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EP97 | Gene names | Senp3, Smt3ip, Smt3ip1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1). | |||||
|
SENP1_HUMAN
|
||||||
| NC score | 0.837669 (rank : 4) | θ value | 7.52953e-39 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP1_MOUSE
|
||||||
| NC score | 0.832687 (rank : 5) | θ value | 7.52953e-39 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP2_MOUSE
|
||||||
| NC score | 0.812624 (rank : 6) | θ value | 2.77131e-33 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZX6, Q544T8, Q9D4Z0 | Gene names | Senp2, Smt3ip2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP2_HUMAN
|
||||||
| NC score | 0.812454 (rank : 7) | θ value | 5.584e-34 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HC62, Q96SR2, Q9P2L5 | Gene names | SENP2, KIAA1331 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SMT3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP6_HUMAN
|
||||||
| NC score | 0.489656 (rank : 8) | θ value | 3.41135e-07 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9GZR1, O94891, Q8TBY4, Q9UJV5 | Gene names | SENP6, KIAA0797, SSP1, SUSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 6 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP6) (SUMO-1-specific protease 1). | |||||
|
SENP7_HUMAN
|
||||||
| NC score | 0.473921 (rank : 9) | θ value | 1.80886e-08 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
|
SENP8_HUMAN
|
||||||
| NC score | 0.296252 (rank : 10) | θ value | 0.000270298 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96LD8, Q96QA4 | Gene names | SENP8, DEN1, NEDP1, PRSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (Protease, cysteine 2) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
SENP8_MOUSE
|
||||||
| NC score | 0.260246 (rank : 11) | θ value | 0.00102713 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D2Z4, Q543P0 | Gene names | Senp8, Den1, Nedp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
RBNS5_HUMAN
|
||||||
| NC score | 0.048953 (rank : 12) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1K0, Q3KP30, Q59EY8, Q8NAQ1 | Gene names | ZFYVE20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rabenosyn-5 (FYVE finger-containing Rab5 effector protein rabenosyn-5) (Zinc finger FYVE domain-containing protein 20) (110 kDa protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.031952 (rank : 13) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.029915 (rank : 14) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.026100 (rank : 15) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.025933 (rank : 16) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.023844 (rank : 17) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
CENPK_MOUSE
|
||||||
| NC score | 0.022812 (rank : 18) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ESN5, Q3UXA9, Q569Q3, Q8C469 | Gene names | Cenpk, Solt | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (SoxLZ/Sox6 leucine zipper-binding protein in testis). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.022440 (rank : 19) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.018649 (rank : 20) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CABYR_MOUSE
|
||||||
| NC score | 0.017974 (rank : 21) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D424, Q91Y41, Q91Y42 | Gene names | Cabyr, Cbp86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86). | |||||
|
LRRC7_HUMAN
|
||||||
| NC score | 0.016545 (rank : 22) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NW7, Q5VXC2, Q5VXC3, Q68D07, Q86VE8, Q8WX20, Q9P2I2 | Gene names | LRRC7, KIAA1365, LAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.016386 (rank : 23) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.015565 (rank : 24) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.015228 (rank : 25) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
TM171_HUMAN
|
||||||
| NC score | 0.015191 (rank : 26) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WVE6, Q8N0S1, Q8TDT7 | Gene names | TMEM171 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 171. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.013750 (rank : 27) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PTHR1_MOUSE
|
||||||
| NC score | 0.013317 (rank : 28) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P41593, Q62119 | Gene names | Pthr1, Pthr | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor (PTH/PTHr receptor) (PTH/PTHrP type I receptor). | |||||
|
CSRP1_HUMAN
|
||||||
| NC score | 0.012642 (rank : 29) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.012640 (rank : 30) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.012632 (rank : 31) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
CSRP1_MOUSE
|
||||||
| NC score | 0.012569 (rank : 32) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
IFI5_MOUSE
|
||||||
| NC score | 0.008932 (rank : 33) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.008843 (rank : 34) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
LRIG1_HUMAN
|
||||||
| NC score | 0.005879 (rank : 35) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JA1, Q6IQ51, Q96CF9, Q9BYB8, Q9UFI4 | Gene names | LRIG1, LIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.005171 (rank : 36) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
K1B24_MOUSE
|
||||||
| NC score | 0.002542 (rank : 37) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61754, Q61755, Q9JM69 | Gene names | Klk1b24, Klk-24, Klk24 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b24 precursor (EC 3.4.21.35) (Glandular kallikrein K24) (Tissue kallikrein 24) (mGK-24). | |||||