Please be patient as the page loads
|
SENP7_HUMAN
|
||||||
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SENP7_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
|
SENP6_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 2) | NC score | 0.771824 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9GZR1, O94891, Q8TBY4, Q9UJV5 | Gene names | SENP6, KIAA0797, SSP1, SUSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 6 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP6) (SUMO-1-specific protease 1). | |||||
|
SENP1_MOUSE
|
||||||
| θ value | 2.28291e-11 (rank : 3) | NC score | 0.518660 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP1_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 4) | NC score | 0.520830 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP2_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 5) | NC score | 0.509018 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HC62, Q96SR2, Q9P2L5 | Gene names | SENP2, KIAA1331 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SMT3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP2_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 6) | NC score | 0.513076 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZX6, Q544T8, Q9D4Z0 | Gene names | Senp2, Smt3ip2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP5_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 7) | NC score | 0.473921 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
SENP3_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 8) | NC score | 0.448861 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |
|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
SENP3_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 9) | NC score | 0.446848 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EP97 | Gene names | Senp3, Smt3ip, Smt3ip1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 10) | NC score | 0.141188 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 11) | NC score | 0.050728 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.098154 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.102355 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.083504 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.092039 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.067147 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.043190 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.089196 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.070788 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.086963 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.070063 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.058121 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.046865 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.075440 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.058467 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.078430 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.062657 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.031216 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.014739 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
K0423_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.041866 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
REST_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.037546 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.017060 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.040044 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.048537 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RPGR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.043952 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
SDC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.040756 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |
|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.069331 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.031445 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.061751 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.064821 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
ZHX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.023229 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.010143 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.050134 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
ESCO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.031674 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
NCA11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.015838 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13591, Q15829, Q16180 | Gene names | NCAM1, NCAM | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 140 kDa isoform precursor (N-CAM 140) (NCAM-140) (CD56 antigen). | |||||
|
NCA12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.015922 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13592, P13593 | Gene names | NCAM1, NCAM | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 120 kDa isoform precursor (N-CAM 120) (NCAM-120) (CD56 antigen). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.050129 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
YSK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.006400 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q56UN5, Q56UN1, Q56UN2, Q56UN3, Q56UN4, Q8N4E9, Q9H5T2 | Gene names | YSK4, RCK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPS1/STE20-related protein kinase YSK4 (EC 2.7.11.1) (Regulated in COPD, protein kinase). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.039564 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.047621 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.030487 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.079161 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.046824 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.033735 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SENP8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.098527 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2Z4, Q543P0 | Gene names | Senp8, Den1, Nedp1 | |||
|
Domain Architecture |
|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
SVIL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.023856 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
VSX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.009681 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
DCR1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.024236 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
KLF6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.000748 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99612, Q5VUT7, Q5VUT8, Q9BT79 | Gene names | KLF6, BCD1, COPEB, CPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 6 (Core promoter element-binding protein) (B- cell-derived protein 1) (Proto-oncogene BCD1) (Transcription factor Zf9) (GC-rich sites-binding factor GBF). | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.019152 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.016553 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
UPAR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.034455 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35456 | Gene names | Plaur | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase plasminogen activator surface receptor, GPI-anchored form precursor (uPAR) (U-PAR) (CD87 antigen). | |||||
|
UPAS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.041044 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35457 | Gene names | Plaur | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase plasminogen activator surface receptor, secreted form precursor (uPAR) (U-PAR) (CD87 antigen). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.010639 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.033728 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
FREM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.011331 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NVD0, Q4W2Q5, Q5H8C0, Q811G9, Q8C4G5, Q8CD46 | Gene names | Frem2, Nv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog) (NV domain-containing protein 1). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.028563 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
HM20A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.030320 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.030146 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
MCM3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.012760 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.027510 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
POT15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.014426 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.028542 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SPAT7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.026716 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VP2, Q8BK23 | Gene names | Spata7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 homolog. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.026640 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
K1383_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.041956 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.015811 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MSH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.013597 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.047308 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
AHTF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.014451 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
CB010_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.036798 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
CJ068_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.023891 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.007308 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.016240 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.021535 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
|
|||||
| Description | Homer protein homolog 1. | |||||
|
IL6RB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.011446 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00560 | Gene names | Il6st | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (CD130 antigen). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.007362 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.008950 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.003734 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
RARG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.008874 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
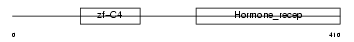
|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.008898 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
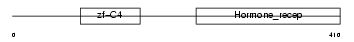
|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.008820 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.008837 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
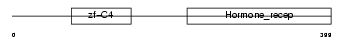
|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RSRC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.024743 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBU6, Q3TF06 | Gene names | Rsrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
SLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.009933 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.028576 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
UBP26_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.011577 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050598 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.050468 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.056760 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.081567 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
SENP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.099131 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96LD8, Q96QA4 | Gene names | SENP8, DEN1, NEDP1, PRSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (Protease, cysteine 2) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
SENP7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9BQF6, Q96PS5, Q9C0F6, Q9HBT5 | Gene names | SENP7, KIAA1707, SSP2, SUSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 7 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP7) (SUMO-1-specific protease 2). | |||||
|
SENP6_HUMAN
|
||||||
| NC score | 0.771824 (rank : 2) | θ value | 1.08223e-53 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9GZR1, O94891, Q8TBY4, Q9UJV5 | Gene names | SENP6, KIAA0797, SSP1, SUSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 6 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP6) (SUMO-1-specific protease 1). | |||||
|
SENP1_HUMAN
|
||||||
| NC score | 0.520830 (rank : 3) | θ value | 5.08577e-11 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP1_MOUSE
|
||||||
| NC score | 0.518660 (rank : 4) | θ value | 2.28291e-11 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SENP2_MOUSE
|
||||||
| NC score | 0.513076 (rank : 5) | θ value | 7.34386e-10 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91ZX6, Q544T8, Q9D4Z0 | Gene names | Senp2, Smt3ip2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP2_HUMAN
|
||||||
| NC score | 0.509018 (rank : 6) | θ value | 7.34386e-10 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HC62, Q96SR2, Q9P2L5 | Gene names | SENP2, KIAA1331 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 2 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP2) (SMT3-specific isopeptidase 2) (Smt3ip2) (Axam2). | |||||
|
SENP5_HUMAN
|
||||||
| NC score | 0.473921 (rank : 7) | θ value | 1.80886e-08 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96HI0, Q96SA5 | Gene names | SENP5 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 5 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP5). | |||||
|
SENP3_HUMAN
|
||||||
| NC score | 0.448861 (rank : 8) | θ value | 3.77169e-06 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4L4, Q96PS4, Q9Y3W9 | Gene names | SENP3, SSP3, SUSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3). | |||||
|
SENP3_MOUSE
|
||||||
| NC score | 0.446848 (rank : 9) | θ value | 3.77169e-06 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EP97 | Gene names | Senp3, Smt3ip, Smt3ip1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 3 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP3) (SUMO-1-specific protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.141188 (rank : 10) | θ value | 5.44631e-05 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.102355 (rank : 11) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SENP8_HUMAN
|
||||||
| NC score | 0.099131 (rank : 12) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96LD8, Q96QA4 | Gene names | SENP8, DEN1, NEDP1, PRSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (Protease, cysteine 2) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
SENP8_MOUSE
|
||||||
| NC score | 0.098527 (rank : 13) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2Z4, Q543P0 | Gene names | Senp8, Den1, Nedp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 8 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP8) (NEDD8-specific protease 1) (Deneddylase-1). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.098154 (rank : 14) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.092039 (rank : 15) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.089196 (rank : 16) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.086963 (rank : 17) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.083504 (rank : 18) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.081567 (rank : 19) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.079161 (rank : 20) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.078430 (rank : 21) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.075440 (rank : 22) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.070788 (rank : 23) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.070063 (rank : 24) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.069331 (rank : 25) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.067147 (rank : 26) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.064821 (rank : 27) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.062657 (rank : 28) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.061751 (rank : 29) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.058467 (rank : 30) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.058121 (rank : 31) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.056760 (rank : 32) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.050728 (rank : 33) | θ value | 0.00665767 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.050598 (rank : 34) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.050468 (rank : 35) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.050134 (rank : 36) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.050129 (rank : 37) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.048537 (rank : 38) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.047621 (rank : 39) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.047308 (rank : 40) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.046865 (rank : 41) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.046824 (rank : 42) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
RPGR_HUMAN
|
||||||
| NC score | 0.043952 (rank : 43) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92834, O00702, O00737, Q8N5T6, Q93039, Q9HD29, Q9UMR1 | Gene names | RPGR, RP3, XLRP3 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.043190 (rank : 44) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
K1383_HUMAN
|
||||||
| NC score | 0.041956 (rank : 45) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.041866 (rank : 46) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
UPAS_MOUSE
|
||||||
| NC score | 0.041044 (rank : 47) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35457 | Gene names | Plaur | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase plasminogen activator surface receptor, secreted form precursor (uPAR) (U-PAR) (CD87 antigen). | |||||
|
SDC3_MOUSE
|
||||||
| NC score | 0.040756 (rank : 48) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64519, Q3UDD9, Q6ZQA4, Q7TQD4 | Gene names | Sdc3, Kiaa0468 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-3 precursor (SYND3). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.040044 (rank : 49) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.039564 (rank : 50) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.037546 (rank : 51) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CB010_HUMAN
|
||||||
| NC score | 0.036798 (rank : 52) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7Z570, Q6ZN26 | Gene names | C2orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf10. | |||||
|
UPAR_MOUSE
|
||||||
| NC score | 0.034455 (rank : 53) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35456 | Gene names | Plaur | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase plasminogen activator surface receptor, GPI-anchored form precursor (uPAR) (U-PAR) (CD87 antigen). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.033735 (rank : 54) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.033728 (rank : 55) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
ESCO1_MOUSE
|
||||||
| NC score | 0.031674 (rank : 56) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z69, Q8BQI2, Q8BR47, Q922F5 | Gene names | Esco1, Kiaa1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.031445 (rank : 57) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.031216 (rank : 58) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.030487 (rank : 59) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
HM20A_HUMAN
|
||||||
| NC score | 0.030320 (rank : 60) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NP66, Q53G31, Q9NSF6 | Gene names | HMG20A, HMGX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (HMG domain-containing protein HMGX1). | |||||
|
HM20A_MOUSE
|
||||||
| NC score | 0.030146 (rank : 61) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DC33, Q3LSF9, Q6NV87, Q8BSK1, Q8C3C1, Q8CAA0, Q9CYG2 | Gene names | Hmg20a, Ibraf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein 20A (HMG box-containing protein 20A) (iBRAF) (Inhibitor of BRAF35) (HMG domain protein HMGX1). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.028576 (rank : 62) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.028563 (rank : 63) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.028542 (rank : 64) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.027510 (rank : 65) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
SPAT7_MOUSE
|
||||||
| NC score | 0.026716 (rank : 66) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VP2, Q8BK23 | Gene names | Spata7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 7 homolog. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.026640 (rank : 67) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RSRC1_MOUSE
|
||||||
| NC score | 0.024743 (rank : 68) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBU6, Q3TF06 | Gene names | Rsrc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine/serine-rich coiled coil protein 1. | |||||
|
DCR1A_MOUSE
|
||||||
| NC score | 0.024236 (rank : 69) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIC3, Q571H5, Q6GQR6 | Gene names | Dclre1a, Kiaa0086, Snm1, Snm1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA cross-link repair 1A protein. | |||||
|
CJ068_HUMAN
|
||||||
| NC score | 0.023891 (rank : 70) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H943, Q8N7T7 | Gene names | C10orf68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf68. | |||||
|
SVIL_HUMAN
|
||||||
| NC score | 0.023856 (rank : 71) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95425, O60611, O60612, Q5VZK5, Q5VZK6, Q9H1R7 | Gene names | SVIL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ZHX1_HUMAN
|
||||||
| NC score | 0.023229 (rank : 72) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.021535 (rank : 73) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.019152 (rank : 74) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.017060 (rank : 75) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.016553 (rank : 76) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.016240 (rank : 77) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
NCA12_HUMAN
|
||||||
| NC score | 0.015922 (rank : 78) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13592, P13593 | Gene names | NCAM1, NCAM | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 120 kDa isoform precursor (N-CAM 120) (NCAM-120) (CD56 antigen). | |||||
|
NCA11_HUMAN
|
||||||
| NC score | 0.015838 (rank : 79) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P13591, Q15829, Q16180 | Gene names | NCAM1, NCAM | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 140 kDa isoform precursor (N-CAM 140) (NCAM-140) (CD56 antigen). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.015811 (rank : 80) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.014739 (rank : 81) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AHTF1_MOUSE
|
||||||
| NC score | 0.014451 (rank : 82) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CJF7, Q8BVJ5, Q8VD55 | Gene names | Ahctf1, Elys | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.014426 (rank : 83) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
MSH4_MOUSE
|
||||||
| NC score | 0.013597 (rank : 84) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
MCM3_HUMAN
|
||||||
| NC score | 0.012760 (rank : 85) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
UBP26_MOUSE
|
||||||
| NC score | 0.011577 (rank : 86) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99MX1 | Gene names | Usp26 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 26 (EC 3.1.2.15) (Ubiquitin thioesterase 26) (Ubiquitin-specific-processing protease 26) (Deubiquitinating enzyme 26). | |||||
|
IL6RB_MOUSE
|
||||||
| NC score | 0.011446 (rank : 87) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00560 | Gene names | Il6st | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-6 receptor beta chain precursor (IL-6R-beta) (Interleukin 6 signal transducer) (Membrane glycoprotein 130) (gp130) (CD130 antigen). | |||||
|
FREM2_MOUSE
|
||||||
| NC score | 0.011331 (rank : 88) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NVD0, Q4W2Q5, Q5H8C0, Q811G9, Q8C4G5, Q8CD46 | Gene names | Frem2, Nv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog) (NV domain-containing protein 1). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.010639 (rank : 89) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.010143 (rank : 90) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.009933 (rank : 91) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
VSX1_MOUSE
|
||||||
| NC score | 0.009681 (rank : 92) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.008950 (rank : 93) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
RARG1_MOUSE
|
||||||
| NC score | 0.008898 (rank : 94) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG1_HUMAN
|
||||||
| NC score | 0.008874 (rank : 95) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG2_MOUSE
|
||||||
| NC score | 0.008837 (rank : 96) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RARG2_HUMAN
|
||||||
| NC score | 0.008820 (rank : 97) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.007362 (rank : 98) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.007308 (rank : 99) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
YSK4_HUMAN
|
||||||
| NC score | 0.006400 (rank : 100) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q56UN5, Q56UN1, Q56UN2, Q56UN3, Q56UN4, Q8N4E9, Q9H5T2 | Gene names | YSK4, RCK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SPS1/STE20-related protein kinase YSK4 (EC 2.7.11.1) (Regulated in COPD, protein kinase). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.003734 (rank : 101) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
KLF6_HUMAN
|
||||||
| NC score | 0.000748 (rank : 102) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99612, Q5VUT7, Q5VUT8, Q9BT79 | Gene names | KLF6, BCD1, COPEB, CPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 6 (Core promoter element-binding protein) (B- cell-derived protein 1) (Proto-oncogene BCD1) (Transcription factor Zf9) (GC-rich sites-binding factor GBF). | |||||