Please be patient as the page loads
|
KCNC3_MOUSE
|
||||||
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995350 (rank : 2) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 173 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 4.84403e-179 (rank : 3) | NC score | 0.990789 (rank : 3) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 6.32649e-179 (rank : 4) | NC score | 0.990686 (rank : 4) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 7.508e-164 (rank : 5) | NC score | 0.987994 (rank : 6) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 2.85304e-163 (rank : 6) | NC score | 0.989055 (rank : 5) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 4.10295e-61 (rank : 7) | NC score | 0.942290 (rank : 7) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 8) | NC score | 0.939084 (rank : 10) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 4.53632e-60 (rank : 9) | NC score | 0.938974 (rank : 11) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 3.84025e-59 (rank : 10) | NC score | 0.936444 (rank : 17) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 3.84025e-59 (rank : 11) | NC score | 0.936838 (rank : 16) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 6.55044e-59 (rank : 12) | NC score | 0.937505 (rank : 14) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 1.90589e-58 (rank : 13) | NC score | 0.937632 (rank : 13) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 7.75577e-52 (rank : 14) | NC score | 0.941521 (rank : 8) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 1.72781e-51 (rank : 15) | NC score | 0.934117 (rank : 19) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 1.72781e-51 (rank : 16) | NC score | 0.934733 (rank : 18) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 2.94719e-51 (rank : 17) | NC score | 0.941188 (rank : 9) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 8.57503e-51 (rank : 18) | NC score | 0.925347 (rank : 27) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 9.48078e-50 (rank : 19) | NC score | 0.932190 (rank : 21) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | 1.23823e-49 (rank : 20) | NC score | 0.925388 (rank : 26) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | 1.73182e-43 (rank : 21) | NC score | 0.921788 (rank : 32) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 22) | NC score | 0.937381 (rank : 15) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 23) | NC score | 0.921851 (rank : 31) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 24) | NC score | 0.922183 (rank : 29) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | 8.59492e-43 (rank : 25) | NC score | 0.921861 (rank : 30) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | 1.91475e-42 (rank : 26) | NC score | 0.920151 (rank : 34) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | 4.2656e-42 (rank : 27) | NC score | 0.920673 (rank : 33) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 28) | NC score | 0.932219 (rank : 20) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 29) | NC score | 0.931643 (rank : 23) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 1.79215e-40 (rank : 30) | NC score | 0.930131 (rank : 24) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 31) | NC score | 0.918021 (rank : 35) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 1.16164e-39 (rank : 32) | NC score | 0.938166 (rank : 12) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 33) | NC score | 0.917472 (rank : 36) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 3.73686e-38 (rank : 34) | NC score | 0.932142 (rank : 22) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 35) | NC score | 0.915031 (rank : 38) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | 8.32485e-38 (rank : 36) | NC score | 0.914776 (rank : 39) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 1.01765e-35 (rank : 37) | NC score | 0.929968 (rank : 25) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 38) | NC score | 0.923268 (rank : 28) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 39) | NC score | 0.915275 (rank : 37) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 40) | NC score | 0.752179 (rank : 40) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 41) | NC score | 0.725138 (rank : 42) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 42) | NC score | 0.724277 (rank : 43) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 43) | NC score | 0.744790 (rank : 41) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 44) | NC score | 0.712925 (rank : 44) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 45) | NC score | 0.706812 (rank : 45) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 46) | NC score | 0.684019 (rank : 47) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 47) | NC score | 0.685269 (rank : 46) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCTD3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 48) | NC score | 0.318386 (rank : 59) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y597, Q49AG7, Q504Q9, Q6PJN6, Q8ND58, Q8NDJ0, Q8WX16 | Gene names | KCTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3 (NY-REN-45 antigen). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 49) | NC score | 0.466932 (rank : 49) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 50) | NC score | 0.467845 (rank : 48) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCD14_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 51) | NC score | 0.269593 (rank : 68) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BQ13 | Gene names | KCTD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD14. | |||||
|
KCTD4_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 52) | NC score | 0.329960 (rank : 55) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD8_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 53) | NC score | 0.190954 (rank : 81) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCTD4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 54) | NC score | 0.327935 (rank : 56) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD5_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 55) | NC score | 0.313280 (rank : 60) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VC57 | Gene names | Kctd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD3_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 56) | NC score | 0.299986 (rank : 64) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
KCNRG_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 57) | NC score | 0.293517 (rank : 66) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q2TUM3, Q6P8J6, Q8C6V1 | Gene names | Kcnrg, Clld4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCD15_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 58) | NC score | 0.334939 (rank : 52) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD5_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 59) | NC score | 0.309611 (rank : 61) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NXV2 | Gene names | KCTD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD8_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 60) | NC score | 0.179665 (rank : 82) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 61) | NC score | 0.170685 (rank : 84) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
KCD15_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 62) | NC score | 0.324683 (rank : 57) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 63) | NC score | 0.318850 (rank : 58) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14681 | Gene names | KCTD2, KIAA0176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 64) | NC score | 0.243055 (rank : 77) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCTD1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 65) | NC score | 0.339988 (rank : 50) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 66) | NC score | 0.339988 (rank : 51) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD6_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 67) | NC score | 0.330346 (rank : 54) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 68) | NC score | 0.333771 (rank : 53) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 69) | NC score | 0.129875 (rank : 105) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 70) | NC score | 0.128960 (rank : 106) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
EP300_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 71) | NC score | 0.024155 (rank : 145) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 72) | NC score | 0.018112 (rank : 151) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
KCNRG_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 73) | NC score | 0.296145 (rank : 65) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N5I3, Q8IU75, Q8N3Q9 | Gene names | KCNRG, CLLD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 74) | NC score | 0.124903 (rank : 107) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 75) | NC score | 0.153071 (rank : 92) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
KCNT1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 76) | NC score | 0.157987 (rank : 89) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
KCTD9_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 77) | NC score | 0.233207 (rank : 78) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 78) | NC score | 0.155417 (rank : 90) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 79) | NC score | 0.037019 (rank : 143) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 0.125558 (rank : 80) | NC score | 0.151518 (rank : 94) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
KCD16_MOUSE
|
||||||
| θ value | 0.163984 (rank : 81) | NC score | 0.163836 (rank : 86) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5DTY9, Q78Y50 | Gene names | Kctd16, Kiaa1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD17_HUMAN
|
||||||
| θ value | 0.163984 (rank : 82) | NC score | 0.252601 (rank : 73) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 83) | NC score | 0.037626 (rank : 142) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
CD248_HUMAN
|
||||||
| θ value | 0.21417 (rank : 84) | NC score | 0.012213 (rank : 158) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.21417 (rank : 85) | NC score | 0.023055 (rank : 147) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
KCD16_HUMAN
|
||||||
| θ value | 0.21417 (rank : 86) | NC score | 0.159063 (rank : 87) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q68DU8, Q9P2M9 | Gene names | KCTD16, KIAA1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 87) | NC score | 0.206518 (rank : 80) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 88) | NC score | 0.258265 (rank : 70) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 89) | NC score | 0.258264 (rank : 71) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 90) | NC score | 0.252754 (rank : 72) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 91) | NC score | 0.251195 (rank : 74) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCTD9_HUMAN
|
||||||
| θ value | 0.21417 (rank : 92) | NC score | 0.227045 (rank : 79) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 0.279714 (rank : 93) | NC score | 0.152126 (rank : 93) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 94) | NC score | 0.247046 (rank : 76) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 95) | NC score | 0.248264 (rank : 75) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 96) | NC score | 0.019833 (rank : 149) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RECQ5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 97) | NC score | 0.016104 (rank : 154) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 98) | NC score | 0.150766 (rank : 95) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 99) | NC score | 0.119350 (rank : 113) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0.365318 (rank : 100) | NC score | 0.113366 (rank : 120) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
KCD12_HUMAN
|
||||||
| θ value | 0.365318 (rank : 101) | NC score | 0.154861 (rank : 91) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCTD7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 102) | NC score | 0.261886 (rank : 69) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCTD7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 103) | NC score | 0.281901 (rank : 67) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCD12_MOUSE
|
||||||
| θ value | 0.62314 (rank : 104) | NC score | 0.148792 (rank : 97) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 105) | NC score | 0.115535 (rank : 115) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 106) | NC score | 0.116003 (rank : 114) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0.813845 (rank : 107) | NC score | 0.112727 (rank : 121) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
E2F4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 108) | NC score | 0.016646 (rank : 152) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
MELPH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 109) | NC score | 0.012925 (rank : 157) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 110) | NC score | 0.023918 (rank : 146) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 111) | NC score | 0.008285 (rank : 167) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 112) | NC score | 0.114527 (rank : 116) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 113) | NC score | 0.014596 (rank : 155) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 114) | NC score | 0.135021 (rank : 102) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 115) | NC score | 0.175936 (rank : 83) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 116) | NC score | 0.004010 (rank : 178) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
PRDM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 117) | NC score | -0.004585 (rank : 193) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75626 | Gene names | PRDM1, BLIMP1 | |||
|
Domain Architecture |
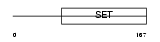
|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1) (Positive regulatory domain I-binding factor 1) (PRDI- binding factor 1) (PRDI-BF1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 1.38821 (rank : 118) | NC score | -0.003242 (rank : 190) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 119) | NC score | 0.006704 (rank : 173) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 120) | NC score | 0.008820 (rank : 166) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
IRK13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 121) | NC score | 0.011655 (rank : 159) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |
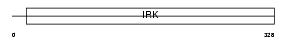
|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 122) | NC score | 0.130772 (rank : 104) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 123) | NC score | 0.113564 (rank : 119) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 124) | NC score | 0.019759 (rank : 150) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 125) | NC score | 0.007637 (rank : 169) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 126) | NC score | 0.002326 (rank : 181) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 127) | NC score | 0.009340 (rank : 165) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 128) | NC score | 0.009801 (rank : 162) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 129) | NC score | 0.165716 (rank : 85) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 130) | NC score | 0.005105 (rank : 177) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 131) | NC score | 0.124845 (rank : 108) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 132) | NC score | 0.123641 (rank : 110) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 3.0926 (rank : 133) | NC score | 0.121572 (rank : 111) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 134) | NC score | 0.016350 (rank : 153) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 135) | NC score | 0.006757 (rank : 172) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DLG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 136) | NC score | -0.000285 (rank : 186) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 3.0926 (rank : 137) | NC score | 0.014528 (rank : 156) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
HXB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 138) | NC score | -0.000083 (rank : 185) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
K2027_HUMAN
|
||||||
| θ value | 3.0926 (rank : 139) | NC score | 0.010050 (rank : 161) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 140) | NC score | 0.142870 (rank : 101) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 141) | NC score | 0.143376 (rank : 100) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 142) | NC score | 0.007525 (rank : 170) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 143) | NC score | 0.005445 (rank : 175) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
SIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 144) | NC score | -0.005383 (rank : 194) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 145) | NC score | 0.009627 (rank : 164) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 146) | NC score | 0.024659 (rank : 144) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 147) | NC score | -0.000323 (rank : 187) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 148) | NC score | 0.300013 (rank : 63) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 149) | NC score | 0.302192 (rank : 62) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 150) | NC score | -0.001393 (rank : 188) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 151) | NC score | 0.067641 (rank : 135) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 152) | NC score | 0.134729 (rank : 103) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 153) | NC score | 0.149624 (rank : 96) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 154) | NC score | 0.000120 (rank : 184) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 155) | NC score | 0.009652 (rank : 163) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 156) | NC score | 0.002741 (rank : 180) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 157) | NC score | -0.003725 (rank : 191) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.106282 (rank : 127) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CF155_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.021889 (rank : 148) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.007863 (rank : 168) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.002229 (rank : 182) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PTPRN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.000541 (rank : 183) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 163) | NC score | -0.004357 (rank : 192) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 164) | NC score | 0.006099 (rank : 174) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 165) | NC score | 0.011409 (rank : 160) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.110432 (rank : 122) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.107204 (rank : 126) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.102375 (rank : 128) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.102305 (rank : 129) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SON_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.005381 (rank : 176) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.007457 (rank : 171) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
TRIM7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | -0.001904 (rank : 189) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.003483 (rank : 179) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.114240 (rank : 117) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.124078 (rank : 109) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.108877 (rank : 125) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.109356 (rank : 123) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.119982 (rank : 112) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
KCD10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.064429 (rank : 140) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H3F6, Q53HN2, Q59FV1, Q6PL47, Q96SU0 | Gene names | KCTD10, ULR061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.067525 (rank : 136) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q922M3 | Gene names | Kctd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCD11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.145201 (rank : 99) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q693B1 | Gene names | KCTD11, REN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCD11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.147630 (rank : 98) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K485 | Gene names | Kctd11, Ren | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.067074 (rank : 137) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ19, Q96P93, Q96SA1 | Gene names | KCTD13, PDIP1, POLDIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1) (TNFAIP1-like protein). | |||||
|
KCD13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.066801 (rank : 138) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BGV7, Q6AXE9 | Gene names | Kctd13, Poldip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1). | |||||
|
KCD18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.100398 (rank : 131) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.079120 (rank : 133) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.113830 (rank : 118) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.066420 (rank : 139) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.070153 (rank : 134) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.100656 (rank : 130) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.099418 (rank : 132) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCTD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.158144 (rank : 88) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CEZ0 | Gene names | Kctd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
SC10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.109154 (rank : 124) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
TNAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.060843 (rank : 141) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13829 | Gene names | TNFAIP1, EDP1 | |||
|
Domain Architecture |
|
|||||
| Description | BTB/POZ domain-containing protein TNFAIP1 (Tumor necrosis factor, alpha-induced protein 1, endothelial) (Protein B12). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 173 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.995350 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 150 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.990789 (rank : 3) | θ value | 4.84403e-179 (rank : 3) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.990686 (rank : 4) | θ value | 6.32649e-179 (rank : 4) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.989055 (rank : 5) | θ value | 2.85304e-163 (rank : 6) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.987994 (rank : 6) | θ value | 7.508e-164 (rank : 5) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.942290 (rank : 7) | θ value | 4.10295e-61 (rank : 7) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.941521 (rank : 8) | θ value | 7.75577e-52 (rank : 14) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.941188 (rank : 9) | θ value | 2.94719e-51 (rank : 17) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.939084 (rank : 10) | θ value | 4.53632e-60 (rank : 8) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.938974 (rank : 11) | θ value | 4.53632e-60 (rank : 9) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.938166 (rank : 12) | θ value | 1.16164e-39 (rank : 32) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.937632 (rank : 13) | θ value | 1.90589e-58 (rank : 13) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.937505 (rank : 14) | θ value | 6.55044e-59 (rank : 12) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.937381 (rank : 15) | θ value | 2.95404e-43 (rank : 22) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.936838 (rank : 16) | θ value | 3.84025e-59 (rank : 11) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.936444 (rank : 17) | θ value | 3.84025e-59 (rank : 10) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 120 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.934733 (rank : 18) | θ value | 1.72781e-51 (rank : 16) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.934117 (rank : 19) | θ value | 1.72781e-51 (rank : 15) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.932219 (rank : 20) | θ value | 2.11701e-41 (rank : 28) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.932190 (rank : 21) | θ value | 9.48078e-50 (rank : 19) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.932142 (rank : 22) | θ value | 3.73686e-38 (rank : 34) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.931643 (rank : 23) | θ value | 4.71619e-41 (rank : 29) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.930131 (rank : 24) | θ value | 1.79215e-40 (rank : 30) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.929968 (rank : 25) | θ value | 1.01765e-35 (rank : 37) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.925388 (rank : 26) | θ value | 1.23823e-49 (rank : 20) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.925347 (rank : 27) | θ value | 8.57503e-51 (rank : 18) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.923268 (rank : 28) | θ value | 1.57365e-28 (rank : 38) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.922183 (rank : 29) | θ value | 6.58091e-43 (rank : 24) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.921861 (rank : 30) | θ value | 8.59492e-43 (rank : 25) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.921851 (rank : 31) | θ value | 3.85808e-43 (rank : 23) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.921788 (rank : 32) | θ value | 1.73182e-43 (rank : 21) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.920673 (rank : 33) | θ value | 4.2656e-42 (rank : 27) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.920151 (rank : 34) | θ value | 1.91475e-42 (rank : 26) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.918021 (rank : 35) | θ value | 8.89434e-40 (rank : 31) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.917472 (rank : 36) | θ value | 2.86122e-38 (rank : 33) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.915275 (rank : 37) | θ value | 8.08199e-25 (rank : 39) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.915031 (rank : 38) | θ value | 4.88048e-38 (rank : 35) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.914776 (rank : 39) | θ value | 8.32485e-38 (rank : 36) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.752179 (rank : 40) | θ value | 4.02038e-16 (rank : 40) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.744790 (rank : 41) | θ value | 5.25075e-16 (rank : 43) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.725138 (rank : 42) | θ value | 4.02038e-16 (rank : 41) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.724277 (rank : 43) | θ value | 4.02038e-16 (rank : 42) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.712925 (rank : 44) | θ value | 5.8054e-15 (rank : 44) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.706812 (rank : 45) | θ value | 4.16044e-13 (rank : 45) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.685269 (rank : 46) | θ value | 5.43371e-13 (rank : 47) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.684019 (rank : 47) | θ value | 5.43371e-13 (rank : 46) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.467845 (rank : 48) | θ value | 0.000121331 (rank : 50) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.466932 (rank : 49) | θ value | 0.000121331 (rank : 49) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCTD1_HUMAN
|
||||||
| NC score | 0.339988 (rank : 50) | θ value | 0.0148317 (rank : 65) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| NC score | 0.339988 (rank : 51) | θ value | 0.0148317 (rank : 66) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCD15_HUMAN
|
||||||
| NC score | 0.334939 (rank : 52) | θ value | 0.00228821 (rank : 58) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD6_MOUSE
|
||||||
| NC score | 0.333771 (rank : 53) | θ value | 0.0148317 (rank : 68) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_HUMAN
|
||||||
| NC score | 0.330346 (rank : 54) | θ value | 0.0148317 (rank : 67) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD4_MOUSE
|
||||||
| NC score | 0.329960 (rank : 55) | θ value | 0.00020696 (rank : 52) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9D7X1, Q8CCQ3, Q9CYK4 | Gene names | Kctd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCTD4_HUMAN
|
||||||
| NC score | 0.327935 (rank : 56) | θ value | 0.000270298 (rank : 54) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8WVF5 | Gene names | KCTD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD4. | |||||
|
KCD15_MOUSE
|
||||||
| NC score | 0.324683 (rank : 57) | θ value | 0.00509761 (rank : 62) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCTD2_HUMAN
|
||||||
| NC score | 0.318850 (rank : 58) | θ value | 0.00665767 (rank : 63) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14681 | Gene names | KCTD2, KIAA0176 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCTD3_HUMAN
|
||||||
| NC score | 0.318386 (rank : 59) | θ value | 2.44474e-05 (rank : 48) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9Y597, Q49AG7, Q504Q9, Q6PJN6, Q8ND58, Q8NDJ0, Q8WX16 | Gene names | KCTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3 (NY-REN-45 antigen). | |||||
|
KCTD5_MOUSE
|
||||||
| NC score | 0.313280 (rank : 60) | θ value | 0.000461057 (rank : 55) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8VC57 | Gene names | Kctd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCTD5_HUMAN
|
||||||
| NC score | 0.309611 (rank : 61) | θ value | 0.00228821 (rank : 59) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NXV2 | Gene names | KCTD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD5. | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.302192 (rank : 62) | θ value | 4.03905 (rank : 149) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.300013 (rank : 63) | θ value | 4.03905 (rank : 148) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCTD3_MOUSE
|
||||||
| NC score | 0.299986 (rank : 64) | θ value | 0.00102713 (rank : 56) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BFX3, Q6P7X4 | Gene names | Kctd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD3. | |||||
|
KCNRG_HUMAN
|
||||||
| NC score | 0.296145 (rank : 65) | θ value | 0.0563607 (rank : 73) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8N5I3, Q8IU75, Q8N3Q9 | Gene names | KCNRG, CLLD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCNRG_MOUSE
|
||||||
| NC score | 0.293517 (rank : 66) | θ value | 0.00175202 (rank : 57) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q2TUM3, Q6P8J6, Q8C6V1 | Gene names | Kcnrg, Clld4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative potassium channel regulatory protein (Potassium channel regulator) (Protein CLLD4). | |||||
|
KCTD7_MOUSE
|
||||||
| NC score | 0.281901 (rank : 67) | θ value | 0.47712 (rank : 103) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8BJK1, Q3USA6, Q8C0S7 | Gene names | Kctd7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCD14_HUMAN
|
||||||
| NC score | 0.269593 (rank : 68) | θ value | 0.00020696 (rank : 51) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BQ13 | Gene names | KCTD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD14. | |||||
|
KCTD7_HUMAN
|
||||||
| NC score | 0.261886 (rank : 69) | θ value | 0.47712 (rank : 102) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96MP8, Q8IVR0 | Gene names | KCTD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD7. | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.258265 (rank : 70) | θ value | 0.21417 (rank : 88) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.258264 (rank : 71) | θ value | 0.21417 (rank : 89) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.252754 (rank : 72) | θ value | 0.21417 (rank : 90) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCD17_HUMAN
|
||||||
| NC score | 0.252601 (rank : 73) | θ value | 0.163984 (rank : 82) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8N5Z5, O95517 | Gene names | KCTD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD17. | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.251195 (rank : 74) | θ value | 0.21417 (rank : 91) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.248264 (rank : 75) | θ value | 0.279714 (rank : 95) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.247046 (rank : 76) | θ value | 0.279714 (rank : 94) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.243055 (rank : 77) | θ value | 0.00869519 (rank : 64) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCTD9_MOUSE
|
||||||
| NC score | 0.233207 (rank : 78) | θ value | 0.0736092 (rank : 77) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80UN1 | Gene names | Kctd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCTD9_HUMAN
|
||||||
| NC score | 0.227045 (rank : 79) | θ value | 0.21417 (rank : 92) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q7L273, Q6NUM8, Q9NXV4 | Gene names | KCTD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD9. | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.206518 (rank : 80) | θ value | 0.21417 (rank : 87) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCTD8_MOUSE
|
||||||
| NC score | 0.190954 (rank : 81) | θ value | 0.00020696 (rank : 53) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q50H33, Q8BR74, Q8C4C2, Q8C906, Q8C9B0, Q8CAA9 | Gene names | Kctd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCTD8_HUMAN
|
||||||
| NC score | 0.179665 (rank : 82) | θ value | 0.00228821 (rank : 60) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZWB6 | Gene names | KCTD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD8. | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.175936 (rank : 83) | θ value | 1.38821 (rank : 115) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.170685 (rank : 84) | θ value | 0.00228821 (rank : 61) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.165716 (rank : 85) | θ value | 2.36792 (rank : 129) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCD16_MOUSE
|
||||||
| NC score | 0.163836 (rank : 86) | θ value | 0.163984 (rank : 81) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5DTY9, Q78Y50 | Gene names | Kctd16, Kiaa1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCD16_HUMAN
|
||||||
| NC score | 0.159063 (rank : 87) | θ value | 0.21417 (rank : 86) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q68DU8, Q9P2M9 | Gene names | KCTD16, KIAA1317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD16. | |||||
|
KCTD2_MOUSE
|
||||||
| NC score | 0.158144 (rank : 88) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CEZ0 | Gene names | Kctd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD2. | |||||
|
KCNT1_MOUSE
|
||||||
| NC score | 0.157987 (rank : 89) | θ value | 0.0736092 (rank : 76) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.155417 (rank : 90) | θ value | 0.0961366 (rank : 78) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
KCD12_HUMAN
|
||||||
| NC score | 0.154861 (rank : 91) | θ value | 0.365318 (rank : 101) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96CX2 | Gene names | KCTD12, C13orf2, KIAA1778, PFET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.153071 (rank : 92) | θ value | 0.0563607 (rank : 75) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.152126 (rank : 93) | θ value | 0.279714 (rank : 93) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.151518 (rank : 94) | θ value | 0.125558 (rank : 80) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.150766 (rank : 95) | θ value | 0.279714 (rank : 98) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.149624 (rank : 96) | θ value | 5.27518 (rank : 153) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCD12_MOUSE
|
||||||
| NC score | 0.148792 (rank : 97) | θ value | 0.62314 (rank : 104) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6WVG3 | Gene names | Kctd12, Pfet1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD12 (Pfetin) (Predominantly fetal expressed T1 domain). | |||||
|
KCD11_MOUSE
|
||||||
| NC score | 0.147630 (rank : 98) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K485 | Gene names | Kctd11, Ren | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCD11_HUMAN
|
||||||
| NC score | 0.145201 (rank : 99) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q693B1 | Gene names | KCTD11, REN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD11. | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.143376 (rank : 100) | θ value | 3.0926 (rank : 141) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.142870 (rank : 101) | θ value | 3.0926 (rank : 140) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.135021 (rank : 102) | θ value | 1.38821 (rank : 114) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.134729 (rank : 103) | θ value | 5.27518 (rank : 152) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.130772 (rank : 104) | θ value | 1.81305 (rank : 122) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.129875 (rank : 105) | θ value | 0.0193708 (rank : 69) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.128960 (rank : 106) | θ value | 0.0330416 (rank : 70) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.124903 (rank : 107) | θ value | 0.0563607 (rank : 74) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.124845 (rank : 108) | θ value | 3.0926 (rank : 131) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.124078 (rank : 109) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.123641 (rank : 110) | θ value | 3.0926 (rank : 132) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.121572 (rank : 111) | θ value | 3.0926 (rank : 133) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.119982 (rank : 112) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.119350 (rank : 113) | θ value | 0.279714 (rank : 99) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.116003 (rank : 114) | θ value | 0.62314 (rank : 106) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.115535 (rank : 115) | θ value | 0.62314 (rank : 105) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.114527 (rank : 116) | θ value | 1.06291 (rank : 112) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.114240 (rank : 117) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.113830 (rank : 118) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.113564 (rank : 119) | θ value | 1.81305 (rank : 123) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.113366 (rank : 120) | θ value | 0.365318 (rank : 100) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.112727 (rank : 121) | θ value | 0.813845 (rank : 107) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.110432 (rank : 122) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.109356 (rank : 123) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.109154 (rank : 124) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.108877 (rank : 125) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.107204 (rank : 126) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.106282 (rank : 127) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.102375 (rank : 128) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.102305 (rank : 129) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.100656 (rank : 130) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCD18_HUMAN
|
||||||
| NC score | 0.100398 (rank : 131) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PI47, Q53T21, Q6NW26, Q6PCD8, Q8N9B7, Q96N73 | Gene names | KCTD18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD18. | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.099418 (rank : 132) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.079120 (rank : 133) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.070153 (rank : 134) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.067641 (rank : 135) | θ value | 5.27518 (rank : 151) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCD10_MOUSE
|
||||||
| NC score | 0.067525 (rank : 136) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q922M3 | Gene names | Kctd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
KCD13_HUMAN
|
||||||
| NC score | 0.067074 (rank : 137) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WZ19, Q96P93, Q96SA1 | Gene names | KCTD13, PDIP1, POLDIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1) (TNFAIP1-like protein). | |||||
|
KCD13_MOUSE
|
||||||
| NC score | 0.066801 (rank : 138) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BGV7, Q6AXE9 | Gene names | Kctd13, Poldip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD13 (Polymerase delta-interacting protein 1). | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.066420 (rank : 139) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCD10_HUMAN
|
||||||
| NC score | 0.064429 (rank : 140) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H3F6, Q53HN2, Q59FV1, Q6PL47, Q96SU0 | Gene names | KCTD10, ULR061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD10. | |||||
|
TNAP1_HUMAN
|
||||||
| NC score | 0.060843 (rank : 141) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13829 | Gene names | TNFAIP1, EDP1 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein TNFAIP1 (Tumor necrosis factor, alpha-induced protein 1, endothelial) (Protein B12). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.037626 (rank : 142) | θ value | 0.163984 (rank : 83) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.037019 (rank : 143) | θ value | 0.0961366 (rank : 79) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.024659 (rank : 144) | θ value | 4.03905 (rank : 146) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.024155 (rank : 145) | θ value | 0.0431538 (rank : 71) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.023918 (rank : 146) | θ value | 1.06291 (rank : 110) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.023055 (rank : 147) | θ value | 0.21417 (rank : 85) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CF155_HUMAN
|
||||||
| NC score | 0.021889 (rank : 148) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.019833 (rank : 149) | θ value | 0.279714 (rank : 96) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.019759 (rank : 150) | θ value | 2.36792 (rank : 124) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.018112 (rank : 151) | θ value | 0.0563607 (rank : 72) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
E2F4_HUMAN
|
||||||
| NC score | 0.016646 (rank : 152) | θ value | 0.813845 (rank : 108) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.016350 (rank : 153) | θ value | 3.0926 (rank : 134) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
RECQ5_HUMAN
|
||||||
| NC score | 0.016104 (rank : 154) | θ value | 0.279714 (rank : 97) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94762, Q9P1W7, Q9UNC8 | Gene names | RECQL5, RECQ5 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q5 (EC 3.6.1.-) (RecQ protein-like 5) (RecQ5). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.014596 (rank : 155) | θ value | 1.38821 (rank : 113) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.014528 (rank : 156) | θ value | 3.0926 (rank : 137) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.012925 (rank : 157) | θ value | 0.813845 (rank : 109) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.012213 (rank : 158) | θ value | 0.21417 (rank : 84) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
IRK13_HUMAN
|
||||||
| NC score | 0.011655 (rank : 159) | θ value | 1.81305 (rank : 121) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.011409 (rank : 160) | θ value | 6.88961 (rank : 165) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.010050 (rank : 161) | θ value | 3.0926 (rank : 139) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.009801 (rank : 162) | θ value | 2.36792 (rank : 128) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.009652 (rank : 163) | θ value | 5.27518 (rank : 155) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.009627 (rank : 164) | θ value | 3.0926 (rank : 145) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.009340 (rank : 165) | θ value | 2.36792 (rank : 127) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.008820 (rank : 166) | θ value | 1.81305 (rank : 120) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.008285 (rank : 167) | θ value | 1.06291 (rank : 111) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.007863 (rank : 168) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.007637 (rank : 169) | θ value | 2.36792 (rank : 125) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.007525 (rank : 170) | θ value | 3.0926 (rank : 142) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.007457 (rank : 171) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.006757 (rank : 172) | θ value | 3.0926 (rank : 135) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.006704 (rank : 173) | θ value | 1.81305 (rank : 119) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.006099 (rank : 174) | θ value | 6.88961 (rank : 164) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.005445 (rank : 175) | θ value | 3.0926 (rank : 143) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.005381 (rank : 176) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.005105 (rank : 177) | θ value | 2.36792 (rank : 130) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.004010 (rank : 178) | θ value | 1.38821 (rank : 116) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.003483 (rank : 179) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.002741 (rank : 180) | θ value | 5.27518 (rank : 156) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.002326 (rank : 181) | θ value | 2.36792 (rank : 126) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.002229 (rank : 182) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PTPRN_MOUSE
|
||||||
| NC score | 0.000541 (rank : 183) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60673, Q62129 | Gene names | Ptprn, Ptp35 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2). | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.000120 (rank : 184) | θ value | 5.27518 (rank : 154) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
HXB3_HUMAN
|
||||||
| NC score | -0.000083 (rank : 185) | θ value | 3.0926 (rank : 138) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14651, O95615, P17484 | Gene names | HOXB3, HOX2G | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2G) (Hox-2.7). | |||||
|
DLG5_HUMAN
|
||||||
| NC score | -0.000285 (rank : 186) | θ value | 3.0926 (rank : 136) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDM6, Q5T1H7, Q5T1H8, Q6DKG3, Q86WC0, Q8TDM7, Q9UE73, Q9Y4E3 | Gene names | DLG5, KIAA0583, PDLG | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg). | |||||
|
HXB3_MOUSE
|
||||||
| NC score | -0.000323 (rank : 187) | θ value | 4.03905 (rank : 147) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | -0.001393 (rank : 188) | θ value | 5.27518 (rank : 150) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
TRIM7_HUMAN
|
||||||
| NC score | -0.001904 (rank : 189) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.003242 (rank : 190) | θ value | 1.38821 (rank : 118) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | -0.003725 (rank : 191) | θ value | 5.27518 (rank : 157) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | -0.004357 (rank : 192) | θ value | 6.88961 (rank : 163) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
PRDM1_HUMAN
|
||||||
| NC score | -0.004585 (rank : 193) | θ value | 1.38821 (rank : 117) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75626 | Gene names | PRDM1, BLIMP1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 1 (PR domain-containing protein 1) (Beta-interferon gene positive-regulatory domain I-binding factor) (BLIMP-1) (Positive regulatory domain I-binding factor 1) (PRDI- binding factor 1) (PRDI-BF1). | |||||
|
SIP1_MOUSE
|
||||||
| NC score | -0.005383 (rank : 194) | θ value | 3.0926 (rank : 144) | |||
| Query Neighborhood Hits | 173 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0G7 | Gene names | Zfhx1b, Sip1, Zfx1b | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 1b (Smad-interacting protein 1). | |||||