Please be patient as the page loads
|
CAC1D_HUMAN
|
||||||
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.946986 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.955621 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.955878 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.951249 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.993159 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.994293 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.959340 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.959422 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.993397 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.993541 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.995599 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.994651 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 7.93343e-89 (rank : 14) | NC score | 0.866604 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 15) | NC score | 0.849178 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 1.22964e-73 (rank : 16) | NC score | 0.846277 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.35953e-72 (rank : 17) | NC score | 0.872910 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 1.35953e-72 (rank : 18) | NC score | 0.847988 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 1.35953e-72 (rank : 19) | NC score | 0.845888 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 5.1662e-72 (rank : 20) | NC score | 0.856105 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 21) | NC score | 0.843867 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 22) | NC score | 0.845320 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 4.37348e-71 (rank : 23) | NC score | 0.842738 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 7.45998e-71 (rank : 24) | NC score | 0.876504 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 1.27248e-70 (rank : 25) | NC score | 0.900685 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 26) | NC score | 0.856943 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 1.07722e-69 (rank : 27) | NC score | 0.864535 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 1.83746e-69 (rank : 28) | NC score | 0.857788 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 3.13423e-69 (rank : 29) | NC score | 0.843201 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 30) | NC score | 0.855685 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 31) | NC score | 0.384016 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 32) | NC score | 0.347207 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 33) | NC score | 0.213791 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 34) | NC score | 0.213903 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 35) | NC score | 0.225777 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 36) | NC score | 0.254690 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 37) | NC score | 0.327781 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 38) | NC score | 0.347683 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 39) | NC score | 0.333459 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 40) | NC score | 0.242233 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 41) | NC score | 0.165063 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 42) | NC score | 0.154226 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 43) | NC score | 0.092886 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 44) | NC score | 0.093876 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 45) | NC score | 0.213601 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 46) | NC score | 0.217154 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 47) | NC score | 0.190012 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 48) | NC score | 0.062902 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.034247 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.170721 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 51) | NC score | 0.118368 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.186314 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.186481 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.014932 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.119591 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.166703 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.180238 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.180396 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.014845 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.022785 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.004172 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
EPC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.009594 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.135797 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.009201 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PLMN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.000123 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20918, Q8CIS2, Q91WJ5 | Gene names | Plg | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
DOCK4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.009492 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.010487 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.127480 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.087686 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.016637 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.056284 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.008432 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
E41LB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.003646 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
GEMI5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.003725 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TEQ6, Q8WWV4, Q969W4, Q9H9K3, Q9UFI5 | Gene names | GEMIN5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.002027 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CD53_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.003600 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61451, Q61721 | Gene names | Cd53 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte surface antigen CD53 (Cell surface glycoprotein CD53). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.140939 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.123041 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.121572 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
LIF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.012841 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09056 | Gene names | Lif | |||
|
Domain Architecture |
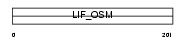
|
|||||
| Description | Leukemia inhibitory factor precursor (LIF) (Differentiation- stimulating factor) (D factor). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.010883 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RPGR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.004901 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.002851 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.005966 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
CENPU_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.006478 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
FRS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.006550 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.139161 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.004622 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PGFRA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | -0.003543 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26618, Q62046 | Gene names | Pdgfra | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
PHAR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.010847 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96KR7, Q707P6, Q9H4T4 | Gene names | PHACTR3, C20orf101, SCAPIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.006980 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.006973 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SEM5A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.000462 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.028453 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
MCLN3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.034806 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | -0.002943 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PAD5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.005687 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
PHAR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.007789 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYK5, Q8BYS8, Q8C058, Q9DB87 | Gene names | Phactr3, Scapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
PPA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.008547 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13686, Q6IAS6 | Gene names | ACP5 | |||
|
Domain Architecture |

|
|||||
| Description | Tartrate-resistant acid phosphatase type 5 precursor (EC 3.1.3.2) (TR- AP) (Tartrate-resistant acid ATPase) (TrATPase) (Acid phosphatase 5, tartrate resistant). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.010340 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
ANR38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.000845 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.016123 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DNL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.025251 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.000898 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
PGFRA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | -0.003833 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16234, Q96KZ7 | Gene names | PDGFRA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.004997 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.002719 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.006185 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | -0.003750 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.002033 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.095321 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | -0.001204 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
S61A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.005162 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9S3 | Gene names | SEC61A2 | |||
|
Domain Architecture |
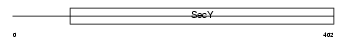
|
|||||
| Description | Protein transport protein Sec61 subunit alpha isoform 2 (Sec61 alpha- 2). | |||||
|
S61A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.005162 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLR1 | Gene names | Sec61a2 | |||
|
Domain Architecture |
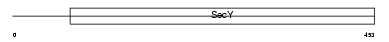
|
|||||
| Description | Protein transport protein Sec61 subunit alpha isoform 2 (Sec61 alpha- 2). | |||||
|
SRCH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.006879 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SV2C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.001875 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.004701 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.036707 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.093159 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.079699 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.078796 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.069408 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.069629 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.076481 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.081410 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.083856 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.090171 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.084035 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.078898 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.063882 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.076542 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.059596 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.055860 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054097 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053083 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.087512 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.063403 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.060758 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.097186 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.068896 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.123751 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
TPTE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.084116 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.995599 (rank : 2) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.994651 (rank : 3) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.994293 (rank : 4) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.993541 (rank : 5) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.993397 (rank : 6) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.993159 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.959422 (rank : 8) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.959340 (rank : 9) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.955878 (rank : 10) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.955621 (rank : 11) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.951249 (rank : 12) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.946986 (rank : 13) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.900685 (rank : 14) | θ value | 1.27248e-70 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.876504 (rank : 15) | θ value | 7.45998e-71 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.872910 (rank : 16) | θ value | 1.35953e-72 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.866604 (rank : 17) | θ value | 7.93343e-89 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.864535 (rank : 18) | θ value | 1.07722e-69 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.857788 (rank : 19) | θ value | 1.83746e-69 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.856943 (rank : 20) | θ value | 4.83544e-70 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.856105 (rank : 21) | θ value | 5.1662e-72 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.855685 (rank : 22) | θ value | 1.36585e-56 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.849178 (rank : 23) | θ value | 3.23591e-74 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.847988 (rank : 24) | θ value | 1.35953e-72 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.846277 (rank : 25) | θ value | 1.22964e-73 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.845888 (rank : 26) | θ value | 1.35953e-72 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.845320 (rank : 27) | θ value | 1.96316e-71 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.843867 (rank : 28) | θ value | 1.96316e-71 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.843201 (rank : 29) | θ value | 3.13423e-69 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.842738 (rank : 30) | θ value | 4.37348e-71 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.384016 (rank : 31) | θ value | 1.38499e-08 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.347683 (rank : 32) | θ value | 9.29e-05 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.347207 (rank : 33) | θ value | 1.53129e-07 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.333459 (rank : 34) | θ value | 9.29e-05 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.327781 (rank : 35) | θ value | 3.19293e-05 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.254690 (rank : 36) | θ value | 8.40245e-06 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.242233 (rank : 37) | θ value | 0.000270298 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.225777 (rank : 38) | θ value | 4.92598e-06 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.217154 (rank : 39) | θ value | 0.00390308 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.213903 (rank : 40) | θ value | 7.59969e-07 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.213791 (rank : 41) | θ value | 7.59969e-07 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.213601 (rank : 42) | θ value | 0.00390308 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.190012 (rank : 43) | θ value | 0.00509761 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.186481 (rank : 44) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.186314 (rank : 45) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.180396 (rank : 46) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.180238 (rank : 47) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.170721 (rank : 48) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.166703 (rank : 49) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.165063 (rank : 50) | θ value | 0.00134147 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.154226 (rank : 51) | θ value | 0.00134147 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.140939 (rank : 52) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.139161 (rank : 53) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.135797 (rank : 54) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.127480 (rank : 55) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.123751 (rank : 56) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.123041 (rank : 57) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.121572 (rank : 58) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.119591 (rank : 59) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.118368 (rank : 60) | θ value | 0.125558 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.097186 (rank : 61) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.095321 (rank : 62) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.093876 (rank : 63) | θ value | 0.00298849 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.093159 (rank : 64) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.092886 (rank : 65) | θ value | 0.00298849 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.090171 (rank : 66) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.087686 (rank : 67) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.087512 (rank : 68) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.084116 (rank : 69) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.084035 (rank : 70) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.083856 (rank : 71) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.081410 (rank : 72) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.079699 (rank : 73) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.078898 (rank : 74) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.078796 (rank : 75) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.076542 (rank : 76) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.076481 (rank : 77) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.069629 (rank : 78) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.069408 (rank : 79) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.068896 (rank : 80) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.063882 (rank : 81) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.063403 (rank : 82) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.062902 (rank : 83) | θ value | 0.0148317 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.060758 (rank : 84) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.059596 (rank : 85) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.056284 (rank : 86) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.055860 (rank : 87) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.054097 (rank : 88) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.053083 (rank : 89) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.036707 (rank : 90) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
MCLN3_MOUSE
|
||||||
| NC score | 0.034806 (rank : 91) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.034247 (rank : 92) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.028453 (rank : 93) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
DNL3_MOUSE
|
||||||
| NC score | 0.025251 (rank : 94) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.022785 (rank : 95) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.016637 (rank : 96) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.016123 (rank : 97) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.014932 (rank : 98) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.014845 (rank : 99) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
LIF_MOUSE
|
||||||
| NC score | 0.012841 (rank : 100) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09056 | Gene names | Lif | |||
|
Domain Architecture |

|
|||||
| Description | Leukemia inhibitory factor precursor (LIF) (Differentiation- stimulating factor) (D factor). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.010883 (rank : 101) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PHAR3_HUMAN
|
||||||
| NC score | 0.010847 (rank : 102) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96KR7, Q707P6, Q9H4T4 | Gene names | PHACTR3, C20orf101, SCAPIN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.010487 (rank : 103) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.010340 (rank : 104) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
EPC2_HUMAN
|
||||||
| NC score | 0.009594 (rank : 105) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
DOCK4_HUMAN
|
||||||
| NC score | 0.009492 (rank : 106) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1I0, O14584, O94824, Q8NB45 | Gene names | DOCK4, KIAA0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.009201 (rank : 107) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PPA5_HUMAN
|
||||||
| NC score | 0.008547 (rank : 108) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13686, Q6IAS6 | Gene names | ACP5 | |||
|
Domain Architecture |

|
|||||
| Description | Tartrate-resistant acid phosphatase type 5 precursor (EC 3.1.3.2) (TR- AP) (Tartrate-resistant acid ATPase) (TrATPase) (Acid phosphatase 5, tartrate resistant). | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.008432 (rank : 109) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
PHAR3_MOUSE
|
||||||
| NC score | 0.007789 (rank : 110) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYK5, Q8BYS8, Q8C058, Q9DB87 | Gene names | Phactr3, Scapin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 3 (Scapinin) (Scaffold-associated PP1- inhibiting protein). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.006980 (rank : 111) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.006973 (rank : 112) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.006879 (rank : 113) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
FRS3_MOUSE
|
||||||
| NC score | 0.006550 (rank : 114) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91WJ0 | Gene names | Frs3, Frs2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor substrate 3 (FGFR substrate 3) (FRS2-beta). | |||||
|
CENPU_HUMAN
|
||||||
| NC score | 0.006478 (rank : 115) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71F23, Q32Q71, Q9H5G1 | Gene names | MLF1IP, CENPU, ICEN24, KLIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein U (CENP-U) (CENP-U(50)) (CENP-50) (Interphase centromere complex protein 24) (MLF1-interacting protein) (KSHV latent nuclear antigen-interacting protein 1). | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.006185 (rank : 116) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.005966 (rank : 117) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
PAD5_MOUSE
|
||||||
| NC score | 0.005687 (rank : 118) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
S61A2_HUMAN
|
||||||
| NC score | 0.005162 (rank : 119) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9S3 | Gene names | SEC61A2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec61 subunit alpha isoform 2 (Sec61 alpha- 2). | |||||
|
S61A2_MOUSE
|
||||||
| NC score | 0.005162 (rank : 120) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLR1 | Gene names | Sec61a2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec61 subunit alpha isoform 2 (Sec61 alpha- 2). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.004997 (rank : 121) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
RPGR_MOUSE
|
||||||
| NC score | 0.004901 (rank : 122) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0X5, O88408, Q9CU92 | Gene names | Rpgr | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator (mRpgr). | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.004701 (rank : 123) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.004622 (rank : 124) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.004172 (rank : 125) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
GEMI5_HUMAN
|
||||||
| NC score | 0.003725 (rank : 126) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TEQ6, Q8WWV4, Q969W4, Q9H9K3, Q9UFI5 | Gene names | GEMIN5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
E41LB_HUMAN
|
||||||
| NC score | 0.003646 (rank : 127) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H329, Q9H328, Q9P2V3 | Gene names | EPB41L4B, EHM2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 4B (Protein EHM2) (FERM-containing protein CG1). | |||||
|
CD53_MOUSE
|
||||||
| NC score | 0.003600 (rank : 128) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61451, Q61721 | Gene names | Cd53 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte surface antigen CD53 (Cell surface glycoprotein CD53). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.002851 (rank : 129) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.002719 (rank : 130) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.002033 (rank : 131) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.002027 (rank : 132) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
SV2C_HUMAN
|
||||||
| NC score | 0.001875 (rank : 133) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q496J9, Q496K1, Q9UPU8 | Gene names | SV2C, KIAA1054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptic vesicle glycoprotein 2C. | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.000898 (rank : 134) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
ANR38_HUMAN
|
||||||
| NC score | 0.000845 (rank : 135) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7N3, Q6P9A0, Q86T71, Q86VE6, Q8NAX3 | Gene names | ANKRD38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 38. | |||||
|
SEM5A_MOUSE
|
||||||
| NC score | 0.000462 (rank : 136) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
PLMN_MOUSE
|
||||||
| NC score | 0.000123 (rank : 137) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20918, Q8CIS2, Q91WJ5 | Gene names | Plg | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
KI21A_MOUSE
|
||||||
| NC score | -0.001204 (rank : 138) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | -0.002943 (rank : 139) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
PGFRA_MOUSE
|
||||||
| NC score | -0.003543 (rank : 140) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26618, Q62046 | Gene names | Pdgfra | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||
|
CP135_MOUSE
|
||||||
| NC score | -0.003750 (rank : 141) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PGFRA_HUMAN
|
||||||
| NC score | -0.003833 (rank : 142) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16234, Q96KZ7 | Gene names | PDGFRA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-alpha) (CD140a antigen). | |||||