Please be patient as the page loads
|
PAD5_MOUSE
|
||||||
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PAD5_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
PAPD5_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996604 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDF8, Q9NW67, Q9Y6C0 | Gene names | PAPD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
POLS_HUMAN
|
||||||
| θ value | 1.80368e-133 (rank : 3) | NC score | 0.968808 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5XG87, O43289, Q9Y6C1 | Gene names | POLS, TRF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7) (Topoisomerase-related function protein 4-1) (TRF4-1) (LAK-1) (DNA polymerase kappa). | |||||
|
POLS_MOUSE
|
||||||
| θ value | 1.52691e-132 (rank : 4) | NC score | 0.953317 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 5) | NC score | 0.396870 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 6) | NC score | 0.385021 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 7) | NC score | 0.316482 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.036869 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
ECHA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.025915 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
MNT_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.028677 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CLSPN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.024213 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
BCL7C_MOUSE
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.022001 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.014283 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
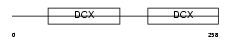
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
LY75_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.011416 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
PAPD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.158790 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0D3, Q3UXJ1, Q8C651 | Gene names | Papd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase) (PAP-associated domain-containing protein 1). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.000326 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
NCTR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.023061 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95944, Q9H562, Q9H563, Q9H564, Q9UMT1, Q9UMT2 | Gene names | NCR2, LY95 | |||
|
Domain Architecture |
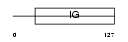
|
|||||
| Description | Natural cytotoxicity triggering receptor 2 precursor (Natural killer cell p44-related protein) (NKp44) (NK-p44) (NK cell-activating receptor) (Lymphocyte antigen 95 homolog) (CD336 antigen). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.018790 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.013204 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.005687 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.012460 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
TNAP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.013811 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.010762 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
NBN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.013712 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.007901 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CDX4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.002833 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | -0.000634 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.008064 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.005260 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.009167 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.003180 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
UNC5C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.004716 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
PAPD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.083434 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVV4, Q659E3, Q6P7E5, Q9HA74 | Gene names | PAPD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (mtPAP) (Polynucleotide adenylyltransferase) (PAP-associated domain- containing protein 1). | |||||
|
SELV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.050299 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
PAD5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
PAPD5_HUMAN
|
||||||
| NC score | 0.996604 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDF8, Q9NW67, Q9Y6C0 | Gene names | PAPD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
POLS_HUMAN
|
||||||
| NC score | 0.968808 (rank : 3) | θ value | 1.80368e-133 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5XG87, O43289, Q9Y6C1 | Gene names | POLS, TRF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7) (Topoisomerase-related function protein 4-1) (TRF4-1) (LAK-1) (DNA polymerase kappa). | |||||
|
POLS_MOUSE
|
||||||
| NC score | 0.953317 (rank : 4) | θ value | 1.52691e-132 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.396870 (rank : 5) | θ value | 2.52405e-10 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.385021 (rank : 6) | θ value | 5.62301e-10 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.316482 (rank : 7) | θ value | 1.09739e-05 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
PAPD1_MOUSE
|
||||||
| NC score | 0.158790 (rank : 8) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0D3, Q3UXJ1, Q8C651 | Gene names | Papd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase) (PAP-associated domain-containing protein 1). | |||||
|
PAPD1_HUMAN
|
||||||
| NC score | 0.083434 (rank : 9) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NVV4, Q659E3, Q6P7E5, Q9HA74 | Gene names | PAPD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (mtPAP) (Polynucleotide adenylyltransferase) (PAP-associated domain- containing protein 1). | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.050299 (rank : 10) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.036869 (rank : 11) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.028677 (rank : 12) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
ECHA_HUMAN
|
||||||
| NC score | 0.025915 (rank : 13) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
CLSPN_HUMAN
|
||||||
| NC score | 0.024213 (rank : 14) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAW4, Q5VYG0, Q6P6H5, Q8IWI1 | Gene names | CLSPN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Claspin (hClaspin) (Hu-Claspin). | |||||
|
NCTR2_HUMAN
|
||||||
| NC score | 0.023061 (rank : 15) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95944, Q9H562, Q9H563, Q9H564, Q9UMT1, Q9UMT2 | Gene names | NCR2, LY95 | |||
|
Domain Architecture |

|
|||||
| Description | Natural cytotoxicity triggering receptor 2 precursor (Natural killer cell p44-related protein) (NKp44) (NK-p44) (NK cell-activating receptor) (Lymphocyte antigen 95 homolog) (CD336 antigen). | |||||
|
BCL7C_MOUSE
|
||||||
| NC score | 0.022001 (rank : 16) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08664, Q99LS2 | Gene names | Bcl7c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member C (B-cell chronic lymphocytic leukemia/lymphoma 7C protein). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.018790 (rank : 17) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.014283 (rank : 18) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
TNAP3_HUMAN
|
||||||
| NC score | 0.013811 (rank : 19) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21580, Q5VXQ7, Q9NSR6 | Gene names | TNFAIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor, alpha-induced protein 3 (EC 3.-.-.-) (Putative DNA-binding protein A20) (Zinc finger protein A20). | |||||
|
NBN_MOUSE
|
||||||
| NC score | 0.013712 (rank : 20) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.013204 (rank : 21) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.012460 (rank : 22) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
LY75_MOUSE
|
||||||
| NC score | 0.011416 (rank : 23) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60767, Q8C7T3, Q91XL8, Q9QUZ6 | Gene names | Ly75, Cd205 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte antigen 75 precursor (DEC-205) (CD205 antigen). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.010762 (rank : 24) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.009167 (rank : 25) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.008064 (rank : 26) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.007901 (rank : 27) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.005687 (rank : 28) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.005260 (rank : 29) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
UNC5C_HUMAN
|
||||||
| NC score | 0.004716 (rank : 30) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.003180 (rank : 31) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CDX4_MOUSE
|
||||||
| NC score | 0.002833 (rank : 32) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.000326 (rank : 33) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
EVI1_HUMAN
|
||||||
| NC score | -0.000634 (rank : 34) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||