Please be patient as the page loads
|
ZCHC6_HUMAN
|
||||||
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCHC6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979667 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCH11_HUMAN
|
||||||
| θ value | 5.01279e-176 (rank : 3) | NC score | 0.862736 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
PAPD1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 4) | NC score | 0.540108 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NVV4, Q659E3, Q6P7E5, Q9HA74 | Gene names | PAPD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (mtPAP) (Polynucleotide adenylyltransferase) (PAP-associated domain- containing protein 1). | |||||
|
PAPD1_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 5) | NC score | 0.550544 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0D3, Q3UXJ1, Q8C651 | Gene names | Papd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase) (PAP-associated domain-containing protein 1). | |||||
|
PAD5_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 6) | NC score | 0.385021 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
PAPD5_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 7) | NC score | 0.387581 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDF8, Q9NW67, Q9Y6C0 | Gene names | PAPD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
POLS_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 8) | NC score | 0.373373 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5XG87, O43289, Q9Y6C1 | Gene names | POLS, TRF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7) (Topoisomerase-related function protein 4-1) (TRF4-1) (LAK-1) (DNA polymerase kappa). | |||||
|
POLS_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 9) | NC score | 0.366589 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 10) | NC score | 0.050948 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.067320 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
CNBP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.169541 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.160027 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.065890 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 15) | NC score | 0.066407 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.068354 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.071916 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.063851 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
K0555_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 19) | NC score | 0.038218 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.023352 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.030279 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
ZFYV9_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.036971 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
PAPOG_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.071919 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BWT3, Q969N1, Q9H8L2, Q9HAD0 | Gene names | PAPOLG, PAP2, PAPG | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme) (Neo- poly(A) polymerase) (Neo-PAP). | |||||
|
PAPOG_MOUSE
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.071497 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.054020 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.128937 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.057062 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
ZCH13_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.152271 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.035492 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
G6PE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.057425 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95479, Q4TT33, Q66I35, Q68DT3 | Gene names | H6PD, GDH | |||
|
Domain Architecture |

|
|||||
| Description | GDH/6PGL endoplasmic bifunctional protein precursor [Includes: Glucose 1-dehydrogenase (EC 1.1.1.47) (Hexose-6-phosphate dehydrogenase); 6- phosphogluconolactonase (EC 3.1.1.31) (6PGL)]. | |||||
|
RB_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.044909 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13405 | Gene names | Rb1, Rb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP105) (RB). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.051039 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.025792 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.047499 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.034122 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
RB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.043869 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06400, P78499, Q5VW46, Q8IZL4 | Gene names | RB1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP110) (P105-RB) (RB). | |||||
|
ZCHC8_HUMAN
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.061942 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.029394 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
WRN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.024874 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.033564 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.050929 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BARD1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.011719 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.046117 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
FOG2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.013464 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.025933 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.026105 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
TAF1L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.046891 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.029124 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
CENB5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.019525 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.019438 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.035722 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.039940 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.039845 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.032373 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.048741 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
AR13B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.010624 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.028968 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.018648 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
BUB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.013167 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
FBX18_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.020317 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2I9, Q8C444, Q99KS0, Q9CTC6, Q9QZG7 | Gene names | Fbxo18, Fbh1, Fbx18 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 18 (EC 3.6.1.-) (F-box DNA helicase 1). | |||||
|
GPDM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.035138 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.013624 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
LRIG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.003040 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JA1, Q6IQ51, Q96CF9, Q9BYB8, Q9UFI4 | Gene names | LRIG1, LIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.027543 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.009449 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.038196 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ZCHC9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.081674 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZN323_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | -0.001073 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96LW9, Q8WWS5 | Gene names | ZNF323 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 323. | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.016605 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
DPOLM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.014445 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIW4, Q9JJW9 | Gene names | Polm | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase mu (EC 2.7.7.7) (Pol Mu). | |||||
|
FA44A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.040178 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
FOG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.011072 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CCH7, Q9Z0F2 | Gene names | Zfpm2, Fog2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (Friend of GATA-2) (FOG-2) (mFOG-2). | |||||
|
GAK18_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.041275 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.044964 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.014403 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
PKHF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.015039 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.035358 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.035341 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.013476 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
ZCRB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.023703 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
CN130_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.017790 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N806, Q86U21, Q86UA9, Q96BY0, Q9NVV6 | Gene names | C14orf130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf130. | |||||
|
CXA8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.006207 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28236 | Gene names | Gja8 | |||
|
Domain Architecture |
|
|||||
| Description | Gap junction alpha-8 protein (Connexin-50) (Cx50) (Lens fiber protein MP70). | |||||
|
GMIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.012099 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
GPDM_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.029852 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
GSUB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.053531 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O96001, Q9UDQ0 | Gene names | GSBS, C7orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-substrate. | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.026039 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
LSP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.025065 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.049541 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.005708 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
OAS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.013125 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29728, Q86XX8 | Gene names | OAS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 2 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 2) (2-5A synthetase 2) (p69 OAS / p71 OAS) (p69OAS / p71OAS). | |||||
|
RBM19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.015336 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.037669 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
TRAT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.045372 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIZ9, Q9NZX5 | Gene names | TRAT1, TCRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor-associated transmembrane adapter 1 (T-cell receptor- interacting molecule) (TRIM) (pp29/30). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.012796 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
ZCHC9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.076443 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R1J3, Q921T6 | Gene names | Zcchc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.009043 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
CD5L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.006915 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.017514 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.018123 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.039080 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.038576 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.042607 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.039482 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.045963 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.038530 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.038473 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.038448 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GMIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.010880 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
NFASC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.004424 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NFASC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.004681 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810U3 | Gene names | Nfasc | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NO40_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.019194 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NP64, Q6PKH4, Q9NYG4, Q9P0M8 | Gene names | ZCCHC17, PS1D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein) (Pnn-interacting nucleolar protein). | |||||
|
PEX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.005539 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
WDR24_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.006308 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFJ9 | Gene names | Wdr24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
ZCHC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.031278 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
|
BC11B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.002329 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0K0, Q9H162 | Gene names | BCL11B, CTIP2, RIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (hRit1) (COUP-TF-interacting protein 2). | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.020344 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.019952 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
CF152_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.016166 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
CH1B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.009621 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LU0, Q3UB77, Q9CXR5 | Gene names | Chmp1b1, Chmp1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-1 (Chromatin-modifying protein 1b-1) (CHMP1b-1). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.009500 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.001245 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.024668 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
HNF3A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.002729 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
LMNB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.007606 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.005992 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.032098 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PLCG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.006383 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
PLCG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.006243 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.007054 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.035186 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.005792 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.016327 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
SFRS7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.025080 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |
|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
SFRS7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.025119 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.010176 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
T2R47_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.002826 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59541, Q645X7 | Gene names | TAS2R47 | |||
|
Domain Architecture |
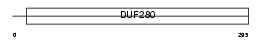
|
|||||
| Description | Taste receptor type 2 member 47 (T2R47). | |||||
|
ZHX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.006348 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.017731 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ZN346_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.013431 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL40, Q68CV9, Q6ZMW1 | Gene names | ZNF346, JAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
ZN403_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.019841 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
ZN638_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.015802 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.005633 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.053568 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCHC6_MOUSE
|
||||||
| NC score | 0.979667 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q5BLK4, Q8CIH3 | Gene names | Zcchc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
ZCH11_HUMAN
|
||||||
| NC score | 0.862736 (rank : 3) | θ value | 5.01279e-176 (rank : 3) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5TAX3, Q12764, Q5TAX4, Q86XZ3 | Gene names | ZCCHC11, KIAA0191 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 11. | |||||
|
PAPD1_MOUSE
|
||||||
| NC score | 0.550544 (rank : 4) | θ value | 4.74913e-17 (rank : 5) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D0D3, Q3UXJ1, Q8C651 | Gene names | Papd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase) (PAP-associated domain-containing protein 1). | |||||
|
PAPD1_HUMAN
|
||||||
| NC score | 0.540108 (rank : 5) | θ value | 2.7842e-17 (rank : 4) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NVV4, Q659E3, Q6P7E5, Q9HA74 | Gene names | PAPD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) RNA polymerase, mitochondrial precursor (EC 2.7.7.19) (PAP) (mtPAP) (Polynucleotide adenylyltransferase) (PAP-associated domain- containing protein 1). | |||||
|
PAPD5_HUMAN
|
||||||
| NC score | 0.387581 (rank : 6) | θ value | 5.62301e-10 (rank : 7) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDF8, Q9NW67, Q9Y6C0 | Gene names | PAPD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
PAD5_MOUSE
|
||||||
| NC score | 0.385021 (rank : 7) | θ value | 5.62301e-10 (rank : 6) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q68ED3, Q8C0K6 | Gene names | Papd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PAP-associated domain-containing protein 5 (EC 2.7.7.-) (Topoisomerase-related function protein 4-2) (TRF4-2). | |||||
|
POLS_HUMAN
|
||||||
| NC score | 0.373373 (rank : 8) | θ value | 3.08544e-08 (rank : 8) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5XG87, O43289, Q9Y6C1 | Gene names | POLS, TRF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7) (Topoisomerase-related function protein 4-1) (TRF4-1) (LAK-1) (DNA polymerase kappa). | |||||
|
POLS_MOUSE
|
||||||
| NC score | 0.366589 (rank : 9) | θ value | 3.08544e-08 (rank : 9) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PB75 | Gene names | Pols | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7). | |||||
|
CNBP_HUMAN
|
||||||
| NC score | 0.169541 (rank : 10) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62633, P20694, Q5QJR0, Q6PJI7, Q96NV3 | Gene names | CNBP, ZNF9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
CNBP_MOUSE
|
||||||
| NC score | 0.160027 (rank : 11) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
ZCH13_HUMAN
|
||||||
| NC score | 0.152271 (rank : 12) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WW36 | Gene names | ZCCHC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 13. | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.128937 (rank : 13) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
ZCHC9_HUMAN
|
||||||
| NC score | 0.081674 (rank : 14) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8N567, Q9H027 | Gene names | ZCCHC9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
ZCHC9_MOUSE
|
||||||
| NC score | 0.076443 (rank : 15) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8R1J3, Q921T6 | Gene names | Zcchc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 9. | |||||
|
PAPOG_HUMAN
|
||||||
| NC score | 0.071919 (rank : 16) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BWT3, Q969N1, Q9H8L2, Q9HAD0 | Gene names | PAPOLG, PAP2, PAPG | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme) (Neo- poly(A) polymerase) (Neo-PAP). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.071916 (rank : 17) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PAPOG_MOUSE
|
||||||
| NC score | 0.071497 (rank : 18) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.068354 (rank : 19) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.067320 (rank : 20) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.066407 (rank : 21) | θ value | 0.0252991 (rank : 15) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.065890 (rank : 22) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.063851 (rank : 23) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
ZCHC8_HUMAN
|
||||||
| NC score | 0.061942 (rank : 24) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NZY4, Q7L2P6, Q8N2K5, Q96SK7, Q9NSS2, Q9NSS3 | Gene names | ZCCHC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
G6PE_HUMAN
|
||||||
| NC score | 0.057425 (rank : 25) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95479, Q4TT33, Q66I35, Q68DT3 | Gene names | H6PD, GDH | |||
|
Domain Architecture |

|
|||||
| Description | GDH/6PGL endoplasmic bifunctional protein precursor [Includes: Glucose 1-dehydrogenase (EC 1.1.1.47) (Hexose-6-phosphate dehydrogenase); 6- phosphogluconolactonase (EC 3.1.1.31) (6PGL)]. | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.057062 (rank : 26) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.054020 (rank : 27) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.053568 (rank : 28) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
GSUB_HUMAN
|
||||||
| NC score | 0.053531 (rank : 29) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O96001, Q9UDQ0 | Gene names | GSBS, C7orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-substrate. | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.051039 (rank : 30) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.050948 (rank : 31) | θ value | 0.00665767 (rank : 10) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.050929 (rank : 32) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.049541 (rank : 33) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.048741 (rank : 34) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.047499 (rank : 35) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
TAF1L_HUMAN
|
||||||
| NC score | 0.046891 (rank : 36) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.046117 (rank : 37) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.045963 (rank : 38) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TRAT1_HUMAN
|
||||||
| NC score | 0.045372 (rank : 39) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIZ9, Q9NZX5 | Gene names | TRAT1, TCRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor-associated transmembrane adapter 1 (T-cell receptor- interacting molecule) (TRIM) (pp29/30). | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.044964 (rank : 40) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RB_MOUSE
|
||||||
| NC score | 0.044909 (rank : 41) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13405 | Gene names | Rb1, Rb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP105) (RB). | |||||
|
RB_HUMAN
|
||||||
| NC score | 0.043869 (rank : 42) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P06400, P78499, Q5VW46, Q8IZL4 | Gene names | RB1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoblastoma-associated protein (PP110) (P105-RB) (RB). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.042607 (rank : 43) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.041275 (rank : 44) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
FA44A_HUMAN
|
||||||
| NC score | 0.040178 (rank : 45) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NFC6, Q6P0M8 | Gene names | FAM44A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM44A. | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.039940 (rank : 46) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.039845 (rank : 47) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.039482 (rank : 48) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.039080 (rank : 49) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.038576 (rank : 50) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.038530 (rank : 51) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.038473 (rank : 52) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.038448 (rank : 53) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.038218 (rank : 54) | θ value | 0.0736092 (rank : 19) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.038196 (rank : 55) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.037669 (rank : 56) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
ZFYV9_HUMAN
|
||||||
| NC score | 0.036971 (rank : 57) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95405, Q9UNE1, Q9Y5R7 | Gene names | ZFYVE9, MADHIP, SARA, SMADIP | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 9 (Mothers against decapentaplegic homolog-interacting protein) (Madh-interacting protein) (Smad anchor for receptor activation) (Receptor activation anchor) (hSARA) (Novel serine protease) (NSP). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.035722 (rank : 58) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.035492 (rank : 59) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.035358 (rank : 60) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.035341 (rank : 61) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.035186 (rank : 62) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
GPDM_MOUSE
|
||||||
| NC score | 0.035138 (rank : 63) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64521, Q61507 | Gene names | Gpd2, Gdm1 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.034122 (rank : 64) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.033564 (rank : 65) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.032373 (rank : 66) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.032098 (rank : 67) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ZCHC5_HUMAN
|
||||||
| NC score | 0.031278 (rank : 68) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8U3 | Gene names | ZCCHC5 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCHC domain-containing protein 5. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.030279 (rank : 69) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
GPDM_HUMAN
|
||||||
| NC score | 0.029852 (rank : 70) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43304, Q59FR1, Q9HAP9 | Gene names | GPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycerol-3-phosphate dehydrogenase, mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M) (mtGPD). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.029394 (rank : 71) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.029124 (rank : 72) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.028968 (rank : 73) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.027543 (rank : 74) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.026105 (rank : 75) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.026039 (rank : 76) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.025933 (rank : 77) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.025792 (rank : 78) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
SFRS7_MOUSE
|
||||||
| NC score | 0.025119 (rank : 79) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BL97, Q8BMC6, Q8BUR2, Q8R2N4, Q8R3E9, Q91YS1 | Gene names | Sfrs7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 7. | |||||
|
SFRS7_HUMAN
|
||||||
| NC score | 0.025080 (rank : 80) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16629 | Gene names | SFRS7 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 7 (Splicing factor 9G8). | |||||
|
LSP1_HUMAN
|
||||||
| NC score | 0.025065 (rank : 81) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P33241, Q16096, Q9BUY8 | Gene names | LSP1, WP34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte-specific protein 1 (Protein pp52) (52 kDa phosphoprotein) (Lymphocyte-specific antigen WP34) (47 kDa actin-binding protein). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.024874 (rank : 82) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.024668 (rank : 83) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
ZCRB1_HUMAN
|
||||||
| NC score | 0.023703 (rank : 84) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TBF4, Q6PJX0 | Gene names | ZCRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (U11/U12 snRNP 31 kDa protein) (U11/U12-31K). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.023352 (rank : 85) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.020344 (rank : 86) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
FBX18_MOUSE
|
||||||
| NC score | 0.020317 (rank : 87) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2I9, Q8C444, Q99KS0, Q9CTC6, Q9QZG7 | Gene names | Fbxo18, Fbh1, Fbx18 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 18 (EC 3.6.1.-) (F-box DNA helicase 1). | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.019952 (rank : 88) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
ZN403_MOUSE
|
||||||
| NC score | 0.019841 (rank : 89) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
CENB5_HUMAN
|
||||||
| NC score | 0.019525 (rank : 90) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96P50, Q9BSR9, Q9C0E7 | Gene names | CENTB5, KIAA1716 | |||
|
Domain Architecture |

|
|||||
| Description | Centaurin-beta 5 (Cnt-b5). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.019438 (rank : 91) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
NO40_HUMAN
|
||||||
| NC score | 0.019194 (rank : 92) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NP64, Q6PKH4, Q9NYG4, Q9P0M8 | Gene names | ZCCHC17, PS1D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein) (Pnn-interacting nucleolar protein). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.018648 (rank : 93) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.018123 (rank : 94) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
CN130_HUMAN
|
||||||
| NC score | 0.017790 (rank : 95) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N806, Q86U21, Q86UA9, Q96BY0, Q9NVV6 | Gene names | C14orf130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf130. | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.017731 (rank : 96) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.017514 (rank : 97) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.016605 (rank : 98) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.016327 (rank : 99) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
CF152_MOUSE
|
||||||
| NC score | 0.016166 (rank : 100) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80ST9, Q9CYM9, Q9D5J9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152 homolog. | |||||
|
ZN638_MOUSE
|
||||||
| NC score | 0.015802 (rank : 101) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61464, Q6DFV9, Q8C941 | Gene names | Znf638, Np220, Zfml | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein). | |||||
|
RBM19_MOUSE
|
||||||
| NC score | 0.015336 (rank : 102) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R3C6, Q8BHR0, Q9CW63 | Gene names | Rbm19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 19 (RNA-binding motif protein 19). | |||||
|
PKHF1_MOUSE
|
||||||
| NC score | 0.015039 (rank : 103) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3TB82, Q99M16 | Gene names | Plekhf1, Lapf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family F member 1 (PH domain- containing family F member 1) (Lysosome-associated apoptosis-inducing protein containing PH and FYVE domains). | |||||
|
DPOLM_MOUSE
|
||||||
| NC score | 0.014445 (rank : 104) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIW4, Q9JJW9 | Gene names | Polm | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase mu (EC 2.7.7.7) (Pol Mu). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.014403 (rank : 105) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.013624 (rank : 106) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.013476 (rank : 107) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
FOG2_HUMAN
|
||||||
| NC score | 0.013464 (rank : 108) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
ZN346_HUMAN
|
||||||
| NC score | 0.013431 (rank : 109) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL40, Q68CV9, Q6ZMW1 | Gene names | ZNF346, JAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
BUB1_HUMAN
|
||||||
| NC score | 0.013167 (rank : 110) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43683, O43430, O43643, O60626 | Gene names | BUB1, BUB1L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 (EC 2.7.11.1) (hBUB1) (BUB1A). | |||||
|
OAS2_HUMAN
|
||||||
| NC score | 0.013125 (rank : 111) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P29728, Q86XX8 | Gene names | OAS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 2 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 2) (2-5A synthetase 2) (p69 OAS / p71 OAS) (p69OAS / p71OAS). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.012796 (rank : 112) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
GMIP_MOUSE
|
||||||
| NC score | 0.012099 (rank : 113) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PGG2, Q6P9S3 | Gene names | Gmip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
BARD1_MOUSE
|
||||||
| NC score | 0.011719 (rank : 114) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
FOG2_MOUSE
|
||||||
| NC score | 0.011072 (rank : 115) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CCH7, Q9Z0F2 | Gene names | Zfpm2, Fog2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (Friend of GATA-2) (FOG-2) (mFOG-2). | |||||
|
GMIP_HUMAN
|
||||||
| NC score | 0.010880 (rank : 116) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P107 | Gene names | GMIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GEM-interacting protein (GMIP). | |||||
|
AR13B_MOUSE
|
||||||
| NC score | 0.010624 (rank : 117) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.010176 (rank : 118) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CH1B1_MOUSE
|
||||||
| NC score | 0.009621 (rank : 119) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LU0, Q3UB77, Q9CXR5 | Gene names | Chmp1b1, Chmp1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 1b-1 (Chromatin-modifying protein 1b-1) (CHMP1b-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.009500 (rank : 120) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.009449 (rank : 121) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.009043 (rank : 122) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
LMNB2_HUMAN
|
||||||
| NC score | 0.007606 (rank : 123) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03252, O75292, Q14734, Q96DF6 | Gene names | LMNB2, LMN2 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B2. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.007054 (rank : 124) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CD5L_MOUSE
|
||||||
| NC score | 0.006915 (rank : 125) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
PLCG1_HUMAN
|
||||||
| NC score | 0.006383 (rank : 126) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
ZHX2_HUMAN
|
||||||
| NC score | 0.006348 (rank : 127) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6X8 | Gene names | ZHX2, AFR1, KIAA0854, RAF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
WDR24_MOUSE
|
||||||
| NC score | 0.006308 (rank : 128) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFJ9 | Gene names | Wdr24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
PLCG1_MOUSE
|
||||||
| NC score | 0.006243 (rank : 129) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
CXA8_MOUSE
|
||||||
| NC score | 0.006207 (rank : 130) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28236 | Gene names | Gja8 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-8 protein (Connexin-50) (Cx50) (Lens fiber protein MP70). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.005992 (rank : 131) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.005792 (rank : 132) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.005708 (rank : 133) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.005633 (rank : 134) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
PEX1_HUMAN
|
||||||
| NC score | 0.005539 (rank : 135) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43933, Q96S71, Q96S72, Q96S73, Q99994 | Gene names | PEX1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome biogenesis factor 1 (Peroxin-1) (Peroxisome biogenesis disorder protein 1). | |||||
|
NFASC_MOUSE
|
||||||
| NC score | 0.004681 (rank : 136) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q810U3 | Gene names | Nfasc | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
NFASC_HUMAN
|
||||||
| NC score | 0.004424 (rank : 137) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94856, Q5T2F0, Q5T2F1, Q5T2F2, Q5T2F3, Q5T2F4, Q5T2F5, Q5T2F6, Q5T2F7, Q5T2F9, Q5T2G0, Q5W9F8, Q68DH3, Q6ZQV6, Q7Z3K1, Q96HT1, Q96K50 | Gene names | NFASC, KIAA0756 | |||
|
Domain Architecture |

|
|||||
| Description | Neurofascin precursor. | |||||
|
LRIG1_HUMAN
|
||||||
| NC score | 0.003040 (rank : 138) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96JA1, Q6IQ51, Q96CF9, Q9BYB8, Q9UFI4 | Gene names | LRIG1, LIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
T2R47_HUMAN
|
||||||
| NC score | 0.002826 (rank : 139) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59541, Q645X7 | Gene names | TAS2R47 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 47 (T2R47). | |||||
|
HNF3A_MOUSE
|
||||||
| NC score | 0.002729 (rank : 140) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35582, Q61108 | Gene names | Foxa1, Hnf3a, Tcf-3a, Tcf3a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 3-alpha (HNF-3A) (Forkhead box protein A1). | |||||
|
BC11B_HUMAN
|
||||||
| NC score | 0.002329 (rank : 141) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0K0, Q9H162 | Gene names | BCL11B, CTIP2, RIT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma/leukemia 11B (B-cell CLL/lymphoma 11B) (Radiation- induced tumor suppressor gene 1 protein) (hRit1) (COUP-TF-interacting protein 2). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.001245 (rank : 142) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
ZN323_HUMAN
|
||||||
| NC score | -0.001073 (rank : 143) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 142 | Target Neighborhood Hits | 802 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96LW9, Q8WWS5 | Gene names | ZNF323 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 323. | |||||