Please be patient as the page loads
|
WWOX_MOUSE
|
||||||
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
WWOX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996602 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
RDH12_MOUSE
|
||||||
| θ value | 9.45883e-58 (rank : 3) | NC score | 0.815223 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
RDH12_HUMAN
|
||||||
| θ value | 1.41344e-53 (rank : 4) | NC score | 0.812596 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
RDH11_MOUSE
|
||||||
| θ value | 2.75849e-49 (rank : 5) | NC score | 0.805910 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYF1, Q3UXM9, Q9D0U5 | Gene names | Rdh11, Arsdr1, Mdt1, Psdr1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (Short-chain aldehyde dehydrogenase) (SCALD) (Cell line MC/9.IL4-derived protein 1) (M42C60). | |||||
|
RDH11_HUMAN
|
||||||
| θ value | 3.60269e-49 (rank : 6) | NC score | 0.803034 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8TC12, Q9NRW0, Q9Y391 | Gene names | RDH11, ARSDR1, PSDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12). | |||||
|
RDH14_HUMAN
|
||||||
| θ value | 1.15895e-47 (rank : 7) | NC score | 0.819358 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HBH5 | Gene names | RDH14, PAN2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RDH14_MOUSE
|
||||||
| θ value | 5.7518e-47 (rank : 8) | NC score | 0.816044 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9ERI6 | Gene names | Rdh14 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RDH13_HUMAN
|
||||||
| θ value | 8.30557e-46 (rank : 9) | NC score | 0.803474 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8NBN7, Q6UX79, Q96G88 | Gene names | RDH13 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
RDH13_MOUSE
|
||||||
| θ value | 1.41672e-45 (rank : 10) | NC score | 0.799093 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CEE7, Q8CC07 | Gene names | Rdh13 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
DHRSX_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 11) | NC score | 0.803492 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N5I4, Q6UWC7, Q8WUS4, Q96GR8, Q9NTF6 | Gene names | DHRSX, CXorf11, DHRS5X | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member on chromosome X precursor (EC 1.1.-.-) (DHRSXY). | |||||
|
DHB7_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 12) | NC score | 0.627110 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56937, Q7Z4V9, Q8WWS2, Q9UF00 | Gene names | HSD17B7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
DHB7_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 13) | NC score | 0.614877 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88736, Q8K5C9, Q921L1 | Gene names | Hsd17b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 14) | NC score | 0.390983 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 15) | NC score | 0.393406 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 16) | NC score | 0.394927 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 17) | NC score | 0.395927 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 18) | NC score | 0.354433 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 19) | NC score | 0.359517 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 20) | NC score | 0.389404 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 21) | NC score | 0.393572 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | 1.74796e-11 (rank : 22) | NC score | 0.454873 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 23) | NC score | 0.290156 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 24) | NC score | 0.353948 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 25) | NC score | 0.295190 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 26) | NC score | 0.356658 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 27) | NC score | 0.367161 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 28) | NC score | 0.360687 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 29) | NC score | 0.373264 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 30) | NC score | 0.368829 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
SAV1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 31) | NC score | 0.446930 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 32) | NC score | 0.445957 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
DHRS8_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 33) | NC score | 0.486711 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8NBQ5, Q96HF6, Q9UKU4 | Gene names | DHRS8, HSD17B11, PAN1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11) (Retinal short-chain dehydrogenase/reductase 2) (retSDR2) (Cutaneous T-cell lymphoma- associated antigen HD-CL-03) (CTCL tumor antigen HD-CL-03). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 34) | NC score | 0.265702 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 35) | NC score | 0.266241 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
DHB13_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 36) | NC score | 0.462401 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
DECR2_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 37) | NC score | 0.391259 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 38) | NC score | 0.233243 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 39) | NC score | 0.346824 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
DECR2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 40) | NC score | 0.359484 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WV68 | Gene names | Decr2, Pdcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2). | |||||
|
DHRS8_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 41) | NC score | 0.471742 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9EQ06, Q3U2P6, Q8BR33, Q8C7S0 | Gene names | Dhrs8, Hsd17b11, Pan1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11). | |||||
|
DHRS4_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 42) | NC score | 0.386661 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
DHB13_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 43) | NC score | 0.449897 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z5P4, Q86W22, Q86W23 | Gene names | HSD17B13, SCDR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 44) | NC score | 0.342441 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
DHRS9_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 45) | NC score | 0.418548 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BPW9, Q5RKX1, Q9NRA9, Q9NRB0 | Gene names | DHRS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (RDH-E2) (Short-chain dehydrogenase/reductase retSDR8) (NADP-dependent retinol dehydrogenase/reductase) (RDHL). | |||||
|
RDH1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 46) | NC score | 0.420050 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92781, Q8TAI2 | Gene names | RDH5, RDH1 | |||
|
Domain Architecture |
|
|||||
| Description | 11-cis retinol dehydrogenase (EC 1.1.1.105) (11-cis RDH). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 47) | NC score | 0.320490 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
DHRS7_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 48) | NC score | 0.478664 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y394, Q9UKU2 | Gene names | DHRS7, RETSDR4 | |||
|
Domain Architecture |
|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4). | |||||
|
PECR_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 49) | NC score | 0.421405 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99MZ7, Q9CX01, Q9JIF4 | Gene names | Pecr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38). | |||||
|
BDH_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 50) | NC score | 0.378543 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q80XN0, Q3UJS9, Q8BK53, Q8R0C8 | Gene names | Bdh1, Bdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
BDH_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 51) | NC score | 0.332292 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02338, Q96ET1, Q9BRZ4 | Gene names | BDH1, BDH | |||
|
Domain Architecture |
|
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 52) | NC score | 0.314322 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
DHRS7_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 53) | NC score | 0.489211 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9CXR1 | Gene names | Dhrs7, Retsdr4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4). | |||||
|
DHI1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 54) | NC score | 0.403469 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P50172 | Gene names | Hsd11b1, Hsd11 | |||
|
Domain Architecture |
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1) (11beta- HSD1A). | |||||
|
DHRS2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 55) | NC score | 0.391439 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13268, Q9H2R2 | Gene names | DHRS2 | |||
|
Domain Architecture |
|
|||||
| Description | Dehydrogenase/reductase SDR family member 2 (EC 1.1.-.-) (HEP27 protein) (Protein D). | |||||
|
PGDH_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 56) | NC score | 0.448661 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCC1, Q61106 | Gene names | Hpgd, Pgdh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
RDH7_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 57) | NC score | 0.422558 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88451 | Gene names | Rdh7, Crad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinol dehydrogenase 7 (EC 1.1.1.105) (Cis-retinol/androgen dehydrogenase type 2) (CRAD-2) (cis-retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme 2). | |||||
|
DHB4_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 58) | NC score | 0.291727 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
DHCA_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 59) | NC score | 0.467951 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P16152 | Gene names | CBR1, CBR, CRN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1) (Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189) (Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin dehydrogenase [NADP+]) (EC 1.1.1.197). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 60) | NC score | 0.330612 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 61) | NC score | 0.334222 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
DHI1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 62) | NC score | 0.428506 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
DHRS4_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 63) | NC score | 0.364518 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LB2, Q9EQU4 | Gene names | Dhrs4, D14Ucla2 | |||
|
Domain Architecture |
|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (mouNRDR). | |||||
|
CBR2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 64) | NC score | 0.414492 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P08074 | Gene names | Cbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Lung carbonyl reductase [NADPH] (EC 1.1.1.184) (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27). | |||||
|
DCXR_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 65) | NC score | 0.387710 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91X52, Q3U5L5, Q9D129, Q9D8W1 | Gene names | Dcxr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase). | |||||
|
DHC3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 66) | NC score | 0.480097 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75828 | Gene names | CBR3 | |||
|
Domain Architecture |
|
|||||
| Description | Carbonyl reductase [NADPH] 3 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 3). | |||||
|
PINL_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 67) | NC score | 0.345149 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
STXB4_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 68) | NC score | 0.165474 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 69) | NC score | 0.167115 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
DHI2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 70) | NC score | 0.345628 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P51661, Q91WK3 | Gene names | Hsd11b2, Hsd11k | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
DHRS9_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 71) | NC score | 0.415330 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q58NB6, Q8BGC7 | Gene names | Dhrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (Short-chain dehydrogenase/reductase retSDR8). | |||||
|
DHCA_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 72) | NC score | 0.452140 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P48758 | Gene names | Cbr1, Cbr | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1). | |||||
|
DHR10_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 73) | NC score | 0.407870 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BPX1, Q9UKU3 | Gene names | DHRS10, SDR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 10 (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase retSDR3). | |||||
|
PGDH_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 74) | NC score | 0.429789 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15428 | Gene names | HPGD, PGDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
DHB12_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 75) | NC score | 0.371164 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q53GQ0, Q96JU2, Q9Y6G8 | Gene names | HSD17B12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR). | |||||
|
DHB4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 76) | NC score | 0.278420 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
DHRS3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 77) | NC score | 0.399633 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |
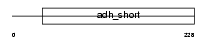
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
BDH2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 78) | NC score | 0.406339 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8JZV9 | Gene names | Bdh2, Dhrs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6). | |||||
|
DHB1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 79) | NC score | 0.351905 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P14061 | Gene names | HSD17B1, E17KSR, EDH17B1, EDHB17 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1) (Placental 17- beta-hydroxysteroid dehydrogenase) (20 alpha-hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHI2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 80) | NC score | 0.276850 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P80365, Q13194, Q8N439, Q96QN8, Q9UC50, Q9UC51 | Gene names | HSD11B2, HSD11K | |||
|
Domain Architecture |
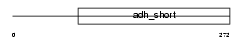
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
DHRS3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 81) | NC score | 0.397831 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |
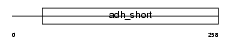
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
DCXR_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 82) | NC score | 0.393989 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z4W1, Q9BTZ3, Q9UHY9 | Gene names | DCXR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase) (Kidney dicarbonyl reductase) (kiDCR) (Carbonyl reductase II) (Sperm surface protein P34H). | |||||
|
GAS7_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 83) | NC score | 0.187892 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 84) | NC score | 0.212043 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
BDH2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 85) | NC score | 0.371816 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BUT1, Q503A0, Q6IA46, Q6UWD3, Q9H8S8, Q9NRX8 | Gene names | BDH2, DHRS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6) (Oxidoreductase UCPA). | |||||
|
PECR_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 86) | NC score | 0.403861 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BY49, Q6IAK9, Q9NRD4, Q9NY60, Q9P1A4 | Gene names | PECR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) (TERP) (HPDHase) (pVI-ARL) (2,4-dienoyl-CoA reductase-related protein) (DCR-RP). | |||||
|
GAS7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 87) | NC score | 0.184867 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
BAG3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 88) | NC score | 0.240514 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
FVT1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 89) | NC score | 0.423564 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q06136 | Gene names | FVT1 | |||
|
Domain Architecture |
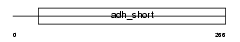
|
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
DHB12_MOUSE
|
||||||
| θ value | 0.21417 (rank : 90) | NC score | 0.348439 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O70503, Q3TID1, Q3U7V9, Q542N3, Q8CI39 | Gene names | Hsd17b12, Kik1 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR) (KIK-I). | |||||
|
DHB3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 91) | NC score | 0.385503 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P37058 | Gene names | HSD17B3, EDH17B3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
DHR11_HUMAN
|
||||||
| θ value | 0.21417 (rank : 92) | NC score | 0.413627 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6UWP2, Q9BUC7, Q9H674 | Gene names | ames=UNQ836/PRO1774 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
FVT1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 93) | NC score | 0.426744 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6GV12 | Gene names | Fvt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
DHR11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 94) | NC score | 0.401370 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q3U0B3, Q5SXB3, Q8R249 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
WBP4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 95) | NC score | 0.101674 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 0.365318 (rank : 96) | NC score | 0.125301 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
BANK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 97) | NC score | 0.042514 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
DHB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 98) | NC score | 0.362186 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P51658, O08898 | Gene names | Hsd17b2, Edh17b2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (17- beta-hydroxysteroid dehydrogenase 2). | |||||
|
PRP40_HUMAN
|
||||||
| θ value | 0.62314 (rank : 99) | NC score | 0.125557 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
DHB1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 100) | NC score | 0.344922 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51656 | Gene names | Hsd17b1, Edh17b1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1). | |||||
|
DRP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 101) | NC score | 0.069949 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
DHB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.331753 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.032027 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.029917 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DECR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.329498 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9CQ62, Q9DCI7 | Gene names | Decr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.081694 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
DECR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.317427 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q16698, Q93085 | Gene names | DECR1, DECR | |||
|
Domain Architecture |
|
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
RHG12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.061978 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.047056 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.046986 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TDPZ3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.003822 (rank : 147) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q717B4 | Gene names | Tdpoz3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 3. | |||||
|
ZCHC6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.013476 (rank : 145) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
DHB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.336215 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P37059 | Gene names | HSD17B2, EDH17B2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (Microsomal 17-beta-hydroxysteroid dehydrogenase) (20 alpha- hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
RT4I1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.014011 (rank : 144) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.026753 (rank : 143) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
HCD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.296831 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.070644 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.007477 (rank : 146) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.057548 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
DHB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.335132 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92506, Q9UIQ1 | Gene names | HSD17B8, FABGL, HKE6, RING2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DHB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.203014 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P50171, Q60958, Q60959, Q9Z1W2 | Gene names | Hsd17b8, H2-Ke6, Hke6 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DHRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.322376 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96LJ7, Q8NDG3, Q96B59, Q96CQ5 | Gene names | DHRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DHRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.348536 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99L04, Q3THW0, Q9D148 | Gene names | Dhrs1, D14ertd484e | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.147564 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.147827 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
HCD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.305462 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.131213 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.212883 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.214089 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.147212 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.149824 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.138340 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.139913 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.172956 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.177887 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.216374 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.216763 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
K0317_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.224412 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.224314 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
SPRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.072525 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35270 | Gene names | SPR | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
SPRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.122870 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64105, Q63996 | Gene names | Spr | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.191733 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.207430 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.204711 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.204928 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.204179 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WWC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.067722 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.996602 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
RDH14_HUMAN
|
||||||
| NC score | 0.819358 (rank : 3) | θ value | 1.15895e-47 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HBH5 | Gene names | RDH14, PAN2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RDH14_MOUSE
|
||||||
| NC score | 0.816044 (rank : 4) | θ value | 5.7518e-47 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9ERI6 | Gene names | Rdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RDH12_MOUSE
|
||||||
| NC score | 0.815223 (rank : 5) | θ value | 9.45883e-58 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
RDH12_HUMAN
|
||||||
| NC score | 0.812596 (rank : 6) | θ value | 1.41344e-53 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
RDH11_MOUSE
|
||||||
| NC score | 0.805910 (rank : 7) | θ value | 2.75849e-49 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QYF1, Q3UXM9, Q9D0U5 | Gene names | Rdh11, Arsdr1, Mdt1, Psdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (Short-chain aldehyde dehydrogenase) (SCALD) (Cell line MC/9.IL4-derived protein 1) (M42C60). | |||||
|
DHRSX_HUMAN
|
||||||
| NC score | 0.803492 (rank : 8) | θ value | 6.15952e-41 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8N5I4, Q6UWC7, Q8WUS4, Q96GR8, Q9NTF6 | Gene names | DHRSX, CXorf11, DHRS5X | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member on chromosome X precursor (EC 1.1.-.-) (DHRSXY). | |||||
|
RDH13_HUMAN
|
||||||
| NC score | 0.803474 (rank : 9) | θ value | 8.30557e-46 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8NBN7, Q6UX79, Q96G88 | Gene names | RDH13 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
RDH11_HUMAN
|
||||||
| NC score | 0.803034 (rank : 10) | θ value | 3.60269e-49 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8TC12, Q9NRW0, Q9Y391 | Gene names | RDH11, ARSDR1, PSDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12). | |||||
|
RDH13_MOUSE
|
||||||
| NC score | 0.799093 (rank : 11) | θ value | 1.41672e-45 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CEE7, Q8CC07 | Gene names | Rdh13 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
DHB7_HUMAN
|
||||||
| NC score | 0.627110 (rank : 12) | θ value | 6.20254e-17 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56937, Q7Z4V9, Q8WWS2, Q9UF00 | Gene names | HSD17B7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
DHB7_MOUSE
|
||||||
| NC score | 0.614877 (rank : 13) | θ value | 6.85773e-16 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88736, Q8K5C9, Q921L1 | Gene names | Hsd17b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
DHRS7_MOUSE
|
||||||
| NC score | 0.489211 (rank : 14) | θ value | 0.000461057 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9CXR1 | Gene names | Dhrs7, Retsdr4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4). | |||||
|
DHRS8_HUMAN
|
||||||
| NC score | 0.486711 (rank : 15) | θ value | 1.17247e-07 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8NBQ5, Q96HF6, Q9UKU4 | Gene names | DHRS8, HSD17B11, PAN1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11) (Retinal short-chain dehydrogenase/reductase 2) (retSDR2) (Cutaneous T-cell lymphoma- associated antigen HD-CL-03) (CTCL tumor antigen HD-CL-03). | |||||
|
DHC3_HUMAN
|
||||||
| NC score | 0.480097 (rank : 16) | θ value | 0.00228821 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O75828 | Gene names | CBR3 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 3 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 3). | |||||
|
DHRS7_HUMAN
|
||||||
| NC score | 0.478664 (rank : 17) | θ value | 0.00020696 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9Y394, Q9UKU2 | Gene names | DHRS7, RETSDR4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4). | |||||
|
DHRS8_MOUSE
|
||||||
| NC score | 0.471742 (rank : 18) | θ value | 1.69304e-06 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9EQ06, Q3U2P6, Q8BR33, Q8C7S0 | Gene names | Dhrs8, Hsd17b11, Pan1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11). | |||||
|
DHCA_HUMAN
|
||||||
| NC score | 0.467951 (rank : 19) | θ value | 0.00102713 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P16152 | Gene names | CBR1, CBR, CRN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1) (Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189) (Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin dehydrogenase [NADP+]) (EC 1.1.1.197). | |||||
|
DHB13_MOUSE
|
||||||
| NC score | 0.462401 (rank : 20) | θ value | 5.81887e-07 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.454873 (rank : 21) | θ value | 1.74796e-11 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
DHCA_MOUSE
|
||||||
| NC score | 0.452140 (rank : 22) | θ value | 0.00665767 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P48758 | Gene names | Cbr1, Cbr | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1). | |||||
|
DHB13_HUMAN
|
||||||
| NC score | 0.449897 (rank : 23) | θ value | 1.43324e-05 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z5P4, Q86W22, Q86W23 | Gene names | HSD17B13, SCDR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9). | |||||
|
PGDH_MOUSE
|
||||||
| NC score | 0.448661 (rank : 24) | θ value | 0.000602161 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCC1, Q61106 | Gene names | Hpgd, Pgdh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
SAV1_HUMAN
|
||||||
| NC score | 0.446930 (rank : 25) | θ value | 2.36244e-08 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| NC score | 0.445957 (rank : 26) | θ value | 3.08544e-08 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
PGDH_HUMAN
|
||||||
| NC score | 0.429789 (rank : 27) | θ value | 0.00665767 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15428 | Gene names | HPGD, PGDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
DHI1_HUMAN
|
||||||
| NC score | 0.428506 (rank : 28) | θ value | 0.00134147 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
FVT1_MOUSE
|
||||||
| NC score | 0.426744 (rank : 29) | θ value | 0.21417 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q6GV12 | Gene names | Fvt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
FVT1_HUMAN
|
||||||
| NC score | 0.423564 (rank : 30) | θ value | 0.163984 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q06136 | Gene names | FVT1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
RDH7_MOUSE
|
||||||
| NC score | 0.422558 (rank : 31) | θ value | 0.000786445 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88451 | Gene names | Rdh7, Crad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinol dehydrogenase 7 (EC 1.1.1.105) (Cis-retinol/androgen dehydrogenase type 2) (CRAD-2) (cis-retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme 2). | |||||
|
PECR_MOUSE
|
||||||
| NC score | 0.421405 (rank : 32) | θ value | 0.00020696 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99MZ7, Q9CX01, Q9JIF4 | Gene names | Pecr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38). | |||||
|
RDH1_HUMAN
|
||||||
| NC score | 0.420050 (rank : 33) | θ value | 7.1131e-05 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92781, Q8TAI2 | Gene names | RDH5, RDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 11-cis retinol dehydrogenase (EC 1.1.1.105) (11-cis RDH). | |||||
|
DHRS9_HUMAN
|
||||||
| NC score | 0.418548 (rank : 34) | θ value | 7.1131e-05 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BPW9, Q5RKX1, Q9NRA9, Q9NRB0 | Gene names | DHRS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (RDH-E2) (Short-chain dehydrogenase/reductase retSDR8) (NADP-dependent retinol dehydrogenase/reductase) (RDHL). | |||||
|
DHRS9_MOUSE
|
||||||
| NC score | 0.415330 (rank : 35) | θ value | 0.00509761 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q58NB6, Q8BGC7 | Gene names | Dhrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (Short-chain dehydrogenase/reductase retSDR8). | |||||
|
CBR2_MOUSE
|
||||||
| NC score | 0.414492 (rank : 36) | θ value | 0.00175202 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P08074 | Gene names | Cbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Lung carbonyl reductase [NADPH] (EC 1.1.1.184) (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27). | |||||
|
DHR11_HUMAN
|
||||||
| NC score | 0.413627 (rank : 37) | θ value | 0.21417 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6UWP2, Q9BUC7, Q9H674 | Gene names | ames=UNQ836/PRO1774 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHR10_HUMAN
|
||||||
| NC score | 0.407870 (rank : 38) | θ value | 0.00665767 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BPX1, Q9UKU3 | Gene names | DHRS10, SDR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 10 (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase retSDR3). | |||||
|
BDH2_MOUSE
|
||||||
| NC score | 0.406339 (rank : 39) | θ value | 0.0252991 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8JZV9 | Gene names | Bdh2, Dhrs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6). | |||||
|
PECR_HUMAN
|
||||||
| NC score | 0.403861 (rank : 40) | θ value | 0.0563607 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BY49, Q6IAK9, Q9NRD4, Q9NY60, Q9P1A4 | Gene names | PECR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) (TERP) (HPDHase) (pVI-ARL) (2,4-dienoyl-CoA reductase-related protein) (DCR-RP). | |||||
|
DHI1_MOUSE
|
||||||
| NC score | 0.403469 (rank : 41) | θ value | 0.000602161 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P50172 | Gene names | Hsd11b1, Hsd11 | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1) (11beta- HSD1A). | |||||
|
DHR11_MOUSE
|
||||||
| NC score | 0.401370 (rank : 42) | θ value | 0.279714 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q3U0B3, Q5SXB3, Q8R249 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHRS3_MOUSE
|
||||||
| NC score | 0.399633 (rank : 43) | θ value | 0.0193708 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
DHRS3_HUMAN
|
||||||
| NC score | 0.397831 (rank : 44) | θ value | 0.0330416 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.395927 (rank : 45) | θ value | 4.16044e-13 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.394927 (rank : 46) | θ value | 4.16044e-13 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
DCXR_HUMAN
|
||||||
| NC score | 0.393989 (rank : 47) | θ value | 0.0431538 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z4W1, Q9BTZ3, Q9UHY9 | Gene names | DCXR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase) (Kidney dicarbonyl reductase) (kiDCR) (Carbonyl reductase II) (Sperm surface protein P34H). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.393572 (rank : 48) | θ value | 1.74796e-11 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.393406 (rank : 49) | θ value | 6.41864e-14 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
DHRS2_HUMAN
|
||||||
| NC score | 0.391439 (rank : 50) | θ value | 0.000602161 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13268, Q9H2R2 | Gene names | DHRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 2 (EC 1.1.-.-) (HEP27 protein) (Protein D). | |||||
|
DECR2_HUMAN
|
||||||
| NC score | 0.391259 (rank : 51) | θ value | 9.92553e-07 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.390983 (rank : 52) | θ value | 6.41864e-14 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.389404 (rank : 53) | θ value | 1.02475e-11 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
DCXR_MOUSE
|
||||||
| NC score | 0.387710 (rank : 54) | θ value | 0.00228821 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91X52, Q3U5L5, Q9D129, Q9D8W1 | Gene names | Dcxr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase). | |||||
|
DHRS4_HUMAN
|
||||||
| NC score | 0.386661 (rank : 55) | θ value | 4.92598e-06 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
DHB3_HUMAN
|
||||||
| NC score | 0.385503 (rank : 56) | θ value | 0.21417 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P37058 | Gene names | HSD17B3, EDH17B3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
BDH_MOUSE
|
||||||
| NC score | 0.378543 (rank : 57) | θ value | 0.000270298 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q80XN0, Q3UJS9, Q8BK53, Q8R0C8 | Gene names | Bdh1, Bdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.373264 (rank : 58) | θ value | 1.25267e-09 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
BDH2_HUMAN
|
||||||
| NC score | 0.371816 (rank : 59) | θ value | 0.0563607 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9BUT1, Q503A0, Q6IA46, Q6UWD3, Q9H8S8, Q9NRX8 | Gene names | BDH2, DHRS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6) (Oxidoreductase UCPA). | |||||
|
DHB12_HUMAN
|
||||||
| NC score | 0.371164 (rank : 60) | θ value | 0.00869519 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q53GQ0, Q96JU2, Q9Y6G8 | Gene names | HSD17B12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.368829 (rank : 61) | θ value | 6.21693e-09 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.367161 (rank : 62) | θ value | 9.59137e-10 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
DHRS4_MOUSE
|
||||||
| NC score | 0.364518 (rank : 63) | θ value | 0.00134147 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LB2, Q9EQU4 | Gene names | Dhrs4, D14Ucla2 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (mouNRDR). | |||||
|
DHB2_MOUSE
|
||||||
| NC score | 0.362186 (rank : 64) | θ value | 0.47712 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P51658, O08898 | Gene names | Hsd17b2, Edh17b2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (17- beta-hydroxysteroid dehydrogenase 2). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.360687 (rank : 65) | θ value | 1.25267e-09 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.359517 (rank : 66) | θ value | 1.02475e-11 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
DECR2_MOUSE
|
||||||
| NC score | 0.359484 (rank : 67) | θ value | 1.69304e-06 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WV68 | Gene names | Decr2, Pdcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.356658 (rank : 68) | θ value | 2.52405e-10 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.354433 (rank : 69) | θ value | 1.02475e-11 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.353948 (rank : 70) | θ value | 3.89403e-11 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
DHB1_HUMAN
|
||||||
| NC score | 0.351905 (rank : 71) | θ value | 0.0252991 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P14061 | Gene names | HSD17B1, E17KSR, EDH17B1, EDHB17 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1) (Placental 17- beta-hydroxysteroid dehydrogenase) (20 alpha-hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHRS1_MOUSE
|
||||||
| NC score | 0.348536 (rank : 72) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99L04, Q3THW0, Q9D148 | Gene names | Dhrs1, D14ertd484e | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DHB12_MOUSE
|
||||||
| NC score | 0.348439 (rank : 73) | θ value | 0.21417 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O70503, Q3TID1, Q3U7V9, Q542N3, Q8CI39 | Gene names | Hsd17b12, Kik1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR) (KIK-I). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.346824 (rank : 74) | θ value | 1.29631e-06 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
DHI2_MOUSE
|
||||||
| NC score | 0.345628 (rank : 75) | θ value | 0.00298849 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P51661, Q91WK3 | Gene names | Hsd11b2, Hsd11k | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
PINL_HUMAN
|
||||||
| NC score | 0.345149 (rank : 76) | θ value | 0.00228821 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
DHB1_MOUSE
|
||||||
| NC score | 0.344922 (rank : 77) | θ value | 1.06291 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51656 | Gene names | Hsd17b1, Edh17b1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.342441 (rank : 78) | θ value | 4.1701e-05 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
DHB2_HUMAN
|
||||||
| NC score | 0.336215 (rank : 79) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P37059 | Gene names | HSD17B2, EDH17B2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (Microsomal 17-beta-hydroxysteroid dehydrogenase) (20 alpha- hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHB8_HUMAN
|
||||||
| NC score | 0.335132 (rank : 80) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92506, Q9UIQ1 | Gene names | HSD17B8, FABGL, HKE6, RING2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.334222 (rank : 81) | θ value | 0.00102713 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
BDH_HUMAN
|
||||||
| NC score | 0.332292 (rank : 82) | θ value | 0.00035302 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02338, Q96ET1, Q9BRZ4 | Gene names | BDH1, BDH | |||
|
Domain Architecture |

|
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
DHB3_MOUSE
|
||||||
| NC score | 0.331753 (rank : 83) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.330612 (rank : 84) | θ value | 0.00102713 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
DECR_MOUSE
|
||||||
| NC score | 0.329498 (rank : 85) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9CQ62, Q9DCI7 | Gene names | Decr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
DHRS1_HUMAN
|
||||||
| NC score | 0.322376 (rank : 86) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96LJ7, Q8NDG3, Q96B59, Q96CQ5 | Gene names | DHRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.320490 (rank : 87) | θ value | 0.000121331 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
DECR_HUMAN
|
||||||
| NC score | 0.317427 (rank : 88) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q16698, Q93085 | Gene names | DECR1, DECR | |||
|
Domain Architecture |

|
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.314322 (rank : 89) | θ value | 0.00035302 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
HCD2_MOUSE
|
||||||
| NC score | 0.305462 (rank : 90) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
HCD2_HUMAN
|
||||||
| NC score | 0.296831 (rank : 91) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.295190 (rank : 92) | θ value | 8.67504e-11 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
DHB4_HUMAN
|
||||||
| NC score | 0.291727 (rank : 93) | θ value | 0.00102713 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.290156 (rank : 94) | θ value | 3.89403e-11 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
DHB4_MOUSE
|
||||||
| NC score | 0.278420 (rank : 95) | θ value | 0.0148317 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
DHI2_HUMAN
|
||||||
| NC score | 0.276850 (rank : 96) | θ value | 0.0330416 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P80365, Q13194, Q8N439, Q96QN8, Q9UC50, Q9UC51 | Gene names | HSD11B2, HSD11K | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.266241 (rank : 97) | θ value | 3.41135e-07 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.265702 (rank : 98) | θ value | 2.61198e-07 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.240514 (rank : 99) | θ value | 0.0961366 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.233243 (rank : 100) | θ value | 1.29631e-06 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.224412 (rank : 101) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.224314 (rank : 102) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.216763 (rank : 103) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.216374 (rank : 104) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.214089 (rank : 105) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.212883 (rank : 106) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.212043 (rank : 107) | θ value | 0.0563607 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.207430 (rank : 108) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.204928 (rank : 109) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.204711 (rank : 110) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.204179 (rank : 111) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
DHB8_MOUSE
|
||||||
| NC score | 0.203014 (rank : 112) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P50171, Q60958, Q60959, Q9Z1W2 | Gene names | Hsd17b8, H2-Ke6, Hke6 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.191733 (rank : 113) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
GAS7_HUMAN
|
||||||
| NC score | 0.187892 (rank : 114) | θ value | 0.0431538 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.184867 (rank : 115) | θ value | 0.0736092 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.177887 (rank : 116) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.172956 (rank : 117) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
STXB4_MOUSE
|
||||||
| NC score | 0.167115 (rank : 118) | θ value | 0.00228821 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.165474 (rank : 119) | θ value | 0.00228821 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.149824 (rank : 120) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.147827 (rank : 121) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.147564 (rank : 122) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.147212 (rank : 123) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.139913 (rank : 124) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.138340 (rank : 125) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.131213 (rank : 126) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
PRP40_HUMAN
|
||||||
| NC score | 0.125557 (rank : 127) | θ value | 0.62314 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O75400, O43856, O75404, Q8TBQ1, Q9H782, Q9NWU9, Q9P0Q2, Q9Y5A8 | Gene names | PRPF40A, FLAF1, FNBP3, HYPA | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Huntingtin yeast partner A) (Huntingtin-interacting protein HYPA/FBP11) (Fas ligand-associated factor 1) (NY-REN-6 antigen). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.125301 (rank : 128) | θ value | 0.365318 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
SPRE_MOUSE
|
||||||
| NC score | 0.122870 (rank : 129) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64105, Q63996 | Gene names | Spr | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
WBP4_MOUSE
|
||||||
| NC score | 0.101674 (rank : 130) | θ value | 0.279714 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61048, Q8K1Z9, Q9CS45 | Gene names | Wbp4, Fbp21, Fnbp21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.081694 (rank : 131) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SPRE_HUMAN
|
||||||
| NC score | 0.072525 (rank : 132) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35270 | Gene names | SPR | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.070644 (rank : 133) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.069949 (rank : 134) | θ value | 1.06291 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
WWC3_HUMAN
|
||||||
| NC score | 0.067722 (rank : 135) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
RHG12_MOUSE
|
||||||
| NC score | 0.061978 (rank : 136) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8C0D4, Q8BVP8 | Gene names | Arhgap12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 12. | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.057548 (rank : 137) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.047056 (rank : 138) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.046986 (rank : 139) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
BANK1_MOUSE
|
||||||
| NC score | 0.042514 (rank : 140) | θ value | 0.47712 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80VH0, Q3U178, Q8BRV6, Q8BRY8 | Gene names | Bank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell scaffold protein with ankyrin repeats (Protein AVIEF). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.032027 (rank : 141) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.029917 (rank : 142) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.026753 (rank : 143) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
RT4I1_HUMAN
|
||||||
| NC score | 0.014011 (rank : 144) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
ZCHC6_HUMAN
|
||||||
| NC score | 0.013476 (rank : 145) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VYS8, Q5H9T0, Q5VYS5, Q5VYS7, Q658Z9, Q659A2, Q6MZJ3, Q8N5F0, Q96N57, Q96NE8, Q9C0F2, Q9H8M6 | Gene names | ZCCHC6, KIAA1711 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 6. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.007477 (rank : 146) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
TDPZ3_MOUSE
|
||||||
| NC score | 0.003822 (rank : 147) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q717B4 | Gene names | Tdpoz3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 3. | |||||