Please be patient as the page loads
|
DHB13_MOUSE
|
||||||
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DHB13_MOUSE
|
||||||
| θ value | 7.23842e-175 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
DHB13_HUMAN
|
||||||
| θ value | 3.89193e-128 (rank : 2) | NC score | 0.983958 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7Z5P4, Q86W22, Q86W23 | Gene names | HSD17B13, SCDR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9). | |||||
|
DHRS8_MOUSE
|
||||||
| θ value | 1.64657e-102 (rank : 3) | NC score | 0.984430 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9EQ06, Q3U2P6, Q8BR33, Q8C7S0 | Gene names | Dhrs8, Hsd17b11, Pan1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11). | |||||
|
DHRS8_HUMAN
|
||||||
| θ value | 2.01279e-100 (rank : 4) | NC score | 0.982766 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NBQ5, Q96HF6, Q9UKU4 | Gene names | DHRS8, HSD17B11, PAN1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11) (Retinal short-chain dehydrogenase/reductase 2) (retSDR2) (Cutaneous T-cell lymphoma- associated antigen HD-CL-03) (CTCL tumor antigen HD-CL-03). | |||||
|
DHRS3_MOUSE
|
||||||
| θ value | 5.03882e-43 (rank : 5) | NC score | 0.921672 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |
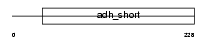
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
DHRS3_HUMAN
|
||||||
| θ value | 4.2656e-42 (rank : 6) | NC score | 0.915819 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |
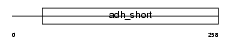
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
DHRS7_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 7) | NC score | 0.836145 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y394, Q9UKU2 | Gene names | DHRS7, RETSDR4 | |||
|
Domain Architecture |
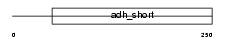
|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4). | |||||
|
BDH2_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 8) | NC score | 0.792598 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8JZV9 | Gene names | Bdh2, Dhrs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6). | |||||
|
DHR11_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 9) | NC score | 0.819973 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q3U0B3, Q5SXB3, Q8R249 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHRS7_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 10) | NC score | 0.823138 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9CXR1 | Gene names | Dhrs7, Retsdr4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4). | |||||
|
DHR11_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 11) | NC score | 0.822062 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6UWP2, Q9BUC7, Q9H674 | Gene names | ames=UNQ836/PRO1774 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
BDH2_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 12) | NC score | 0.788756 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BUT1, Q503A0, Q6IA46, Q6UWD3, Q9H8S8, Q9NRX8 | Gene names | BDH2, DHRS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6) (Oxidoreductase UCPA). | |||||
|
DHI1_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 13) | NC score | 0.800966 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P50172 | Gene names | Hsd11b1, Hsd11 | |||
|
Domain Architecture |
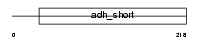
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1) (11beta- HSD1A). | |||||
|
DHI1_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 14) | NC score | 0.814779 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
DHRS4_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 15) | NC score | 0.773590 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
DHRS4_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 16) | NC score | 0.749386 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LB2, Q9EQU4 | Gene names | Dhrs4, D14Ucla2 | |||
|
Domain Architecture |
|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (mouNRDR). | |||||
|
FVT1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 17) | NC score | 0.779552 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6GV12 | Gene names | Fvt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
FVT1_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 18) | NC score | 0.775671 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q06136 | Gene names | FVT1 | |||
|
Domain Architecture |
|
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
PGDH_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 19) | NC score | 0.769563 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15428 | Gene names | HPGD, PGDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
CBR2_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 20) | NC score | 0.734020 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P08074 | Gene names | Cbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Lung carbonyl reductase [NADPH] (EC 1.1.1.184) (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27). | |||||
|
DHR10_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 21) | NC score | 0.746430 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BPX1, Q9UKU3 | Gene names | DHRS10, SDR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 10 (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase retSDR3). | |||||
|
RDH12_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 22) | NC score | 0.622245 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
PGDH_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 23) | NC score | 0.765576 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCC1, Q61106 | Gene names | Hpgd, Pgdh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
DHB12_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 24) | NC score | 0.748833 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q53GQ0, Q96JU2, Q9Y6G8 | Gene names | HSD17B12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR). | |||||
|
DHB3_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 25) | NC score | 0.725615 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P37058 | Gene names | HSD17B3, EDH17B3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
RDH7_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 26) | NC score | 0.660666 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88451 | Gene names | Rdh7, Crad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinol dehydrogenase 7 (EC 1.1.1.105) (Cis-retinol/androgen dehydrogenase type 2) (CRAD-2) (cis-retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme 2). | |||||
|
DHB1_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 27) | NC score | 0.708657 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51656 | Gene names | Hsd17b1, Edh17b1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1). | |||||
|
DHB1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 28) | NC score | 0.729303 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P14061 | Gene names | HSD17B1, E17KSR, EDH17B1, EDHB17 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1) (Placental 17- beta-hydroxysteroid dehydrogenase) (20 alpha-hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
RDH13_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 29) | NC score | 0.681284 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CEE7, Q8CC07 | Gene names | Rdh13 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
DCXR_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 30) | NC score | 0.716906 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z4W1, Q9BTZ3, Q9UHY9 | Gene names | DCXR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase) (Kidney dicarbonyl reductase) (kiDCR) (Carbonyl reductase II) (Sperm surface protein P34H). | |||||
|
DHB12_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 31) | NC score | 0.740261 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70503, Q3TID1, Q3U7V9, Q542N3, Q8CI39 | Gene names | Hsd17b12, Kik1 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR) (KIK-I). | |||||
|
RDH13_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 32) | NC score | 0.672749 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NBN7, Q6UX79, Q96G88 | Gene names | RDH13 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
DHI2_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 33) | NC score | 0.607012 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51661, Q91WK3 | Gene names | Hsd11b2, Hsd11k | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
DHRS9_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 34) | NC score | 0.637124 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BPW9, Q5RKX1, Q9NRA9, Q9NRB0 | Gene names | DHRS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (RDH-E2) (Short-chain dehydrogenase/reductase retSDR8) (NADP-dependent retinol dehydrogenase/reductase) (RDHL). | |||||
|
DHRS9_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 35) | NC score | 0.650218 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q58NB6, Q8BGC7 | Gene names | Dhrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (Short-chain dehydrogenase/reductase retSDR8). | |||||
|
DHB4_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 36) | NC score | 0.615694 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
RDH1_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 37) | NC score | 0.655148 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92781, Q8TAI2 | Gene names | RDH5, RDH1 | |||
|
Domain Architecture |
|
|||||
| Description | 11-cis retinol dehydrogenase (EC 1.1.1.105) (11-cis RDH). | |||||
|
DHB3_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 38) | NC score | 0.685568 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
BDH_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 39) | NC score | 0.647446 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80XN0, Q3UJS9, Q8BK53, Q8R0C8 | Gene names | Bdh1, Bdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
DCXR_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 40) | NC score | 0.703297 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91X52, Q3U5L5, Q9D129, Q9D8W1 | Gene names | Dcxr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase). | |||||
|
DHB4_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 41) | NC score | 0.605510 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
DHRS2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 42) | NC score | 0.748108 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13268, Q9H2R2 | Gene names | DHRS2 | |||
|
Domain Architecture |
|
|||||
| Description | Dehydrogenase/reductase SDR family member 2 (EC 1.1.-.-) (HEP27 protein) (Protein D). | |||||
|
RDH12_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 43) | NC score | 0.610169 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
RDH14_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 44) | NC score | 0.575203 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HBH5 | Gene names | RDH14, PAN2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
DHB8_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 45) | NC score | 0.676613 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92506, Q9UIQ1 | Gene names | HSD17B8, FABGL, HKE6, RING2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
HCD2_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 46) | NC score | 0.631122 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
RDH14_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 47) | NC score | 0.574118 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9ERI6 | Gene names | Rdh14 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
BDH_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 48) | NC score | 0.605408 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02338, Q96ET1, Q9BRZ4 | Gene names | BDH1, BDH | |||
|
Domain Architecture |
|
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
PECR_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 49) | NC score | 0.673604 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BY49, Q6IAK9, Q9NRD4, Q9NY60, Q9P1A4 | Gene names | PECR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) (TERP) (HPDHase) (pVI-ARL) (2,4-dienoyl-CoA reductase-related protein) (DCR-RP). | |||||
|
RDH11_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 50) | NC score | 0.632746 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8TC12, Q9NRW0, Q9Y391 | Gene names | RDH11, ARSDR1, PSDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12). | |||||
|
RDH11_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 51) | NC score | 0.620362 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QYF1, Q3UXM9, Q9D0U5 | Gene names | Rdh11, Arsdr1, Mdt1, Psdr1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (Short-chain aldehyde dehydrogenase) (SCALD) (Cell line MC/9.IL4-derived protein 1) (M42C60). | |||||
|
DHB2_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 52) | NC score | 0.553406 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P37059 | Gene names | HSD17B2, EDH17B2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (Microsomal 17-beta-hydroxysteroid dehydrogenase) (20 alpha- hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 53) | NC score | 0.462401 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
DECR_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 54) | NC score | 0.564080 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q16698, Q93085 | Gene names | DECR1, DECR | |||
|
Domain Architecture |
|
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 55) | NC score | 0.437976 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
PECR_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 56) | NC score | 0.635010 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99MZ7, Q9CX01, Q9JIF4 | Gene names | Pecr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38). | |||||
|
DHB8_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 57) | NC score | 0.588451 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P50171, Q60958, Q60959, Q9Z1W2 | Gene names | Hsd17b8, H2-Ke6, Hke6 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DHI2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 58) | NC score | 0.551370 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P80365, Q13194, Q8N439, Q96QN8, Q9UC50, Q9UC51 | Gene names | HSD11B2, HSD11K | |||
|
Domain Architecture |
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
DHRS1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 59) | NC score | 0.625935 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96LJ7, Q8NDG3, Q96B59, Q96CQ5 | Gene names | DHRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DHRS1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 60) | NC score | 0.660656 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99L04, Q3THW0, Q9D148 | Gene names | Dhrs1, D14ertd484e | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DECR_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 61) | NC score | 0.577227 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9CQ62, Q9DCI7 | Gene names | Decr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
DHRSX_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 62) | NC score | 0.569120 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N5I4, Q6UWC7, Q8WUS4, Q96GR8, Q9NTF6 | Gene names | DHRSX, CXorf11, DHRS5X | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member on chromosome X precursor (EC 1.1.-.-) (DHRSXY). | |||||
|
DHC3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 63) | NC score | 0.491639 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75828 | Gene names | CBR3 | |||
|
Domain Architecture |
|
|||||
| Description | Carbonyl reductase [NADPH] 3 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 3). | |||||
|
DHB7_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 64) | NC score | 0.317752 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88736, Q8K5C9, Q921L1 | Gene names | Hsd17b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
DHB7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 65) | NC score | 0.349217 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56937, Q7Z4V9, Q8WWS2, Q9UF00 | Gene names | HSD17B7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
DHCA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.453575 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P16152 | Gene names | CBR1, CBR, CRN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1) (Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189) (Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin dehydrogenase [NADP+]) (EC 1.1.1.197). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.025155 (rank : 76) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.022554 (rank : 77) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
RT4I1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.016990 (rank : 78) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||
|
CT079_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.061289 (rank : 75) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJQ7 | Gene names | C20orf79 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf79. | |||||
|
CT079_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.061306 (rank : 74) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DAH1 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf79 homolog. | |||||
|
DECR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.496773 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
DECR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.478583 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WV68 | Gene names | Decr2, Pdcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2). | |||||
|
DHB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.609721 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P51658, O08898 | Gene names | Hsd17b2, Edh17b2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (17- beta-hydroxysteroid dehydrogenase 2). | |||||
|
DHCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.439847 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P48758 | Gene names | Cbr1, Cbr | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1). | |||||
|
HCD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.515241 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
SPRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.209065 (rank : 73) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35270 | Gene names | SPR | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
SPRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.244182 (rank : 72) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64105, Q63996 | Gene names | Spr | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
DHB13_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.23842e-175 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
DHRS8_MOUSE
|
||||||
| NC score | 0.984430 (rank : 2) | θ value | 1.64657e-102 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9EQ06, Q3U2P6, Q8BR33, Q8C7S0 | Gene names | Dhrs8, Hsd17b11, Pan1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11). | |||||
|
DHB13_HUMAN
|
||||||
| NC score | 0.983958 (rank : 3) | θ value | 3.89193e-128 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7Z5P4, Q86W22, Q86W23 | Gene names | HSD17B13, SCDR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9). | |||||
|
DHRS8_HUMAN
|
||||||
| NC score | 0.982766 (rank : 4) | θ value | 2.01279e-100 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NBQ5, Q96HF6, Q9UKU4 | Gene names | DHRS8, HSD17B11, PAN1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11) (Retinal short-chain dehydrogenase/reductase 2) (retSDR2) (Cutaneous T-cell lymphoma- associated antigen HD-CL-03) (CTCL tumor antigen HD-CL-03). | |||||
|
DHRS3_MOUSE
|
||||||
| NC score | 0.921672 (rank : 5) | θ value | 5.03882e-43 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
DHRS3_HUMAN
|
||||||
| NC score | 0.915819 (rank : 6) | θ value | 4.2656e-42 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
DHRS7_HUMAN
|
||||||
| NC score | 0.836145 (rank : 7) | θ value | 2.5182e-18 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y394, Q9UKU2 | Gene names | DHRS7, RETSDR4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4). | |||||
|
DHRS7_MOUSE
|
||||||
| NC score | 0.823138 (rank : 8) | θ value | 1.058e-16 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9CXR1 | Gene names | Dhrs7, Retsdr4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4). | |||||
|
DHR11_HUMAN
|
||||||
| NC score | 0.822062 (rank : 9) | θ value | 1.38178e-16 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6UWP2, Q9BUC7, Q9H674 | Gene names | ames=UNQ836/PRO1774 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHR11_MOUSE
|
||||||
| NC score | 0.819973 (rank : 10) | θ value | 1.058e-16 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q3U0B3, Q5SXB3, Q8R249 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHI1_HUMAN
|
||||||
| NC score | 0.814779 (rank : 11) | θ value | 7.58209e-15 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
DHI1_MOUSE
|
||||||
| NC score | 0.800966 (rank : 12) | θ value | 1.52774e-15 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P50172 | Gene names | Hsd11b1, Hsd11 | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1) (11beta- HSD1A). | |||||
|
BDH2_MOUSE
|
||||||
| NC score | 0.792598 (rank : 13) | θ value | 8.10077e-17 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8JZV9 | Gene names | Bdh2, Dhrs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6). | |||||
|
BDH2_HUMAN
|
||||||
| NC score | 0.788756 (rank : 14) | θ value | 4.02038e-16 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9BUT1, Q503A0, Q6IA46, Q6UWD3, Q9H8S8, Q9NRX8 | Gene names | BDH2, DHRS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6) (Oxidoreductase UCPA). | |||||
|
FVT1_MOUSE
|
||||||
| NC score | 0.779552 (rank : 15) | θ value | 2.69671e-12 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q6GV12 | Gene names | Fvt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
FVT1_HUMAN
|
||||||
| NC score | 0.775671 (rank : 16) | θ value | 1.33837e-11 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q06136 | Gene names | FVT1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
DHRS4_HUMAN
|
||||||
| NC score | 0.773590 (rank : 17) | θ value | 3.18553e-13 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
PGDH_HUMAN
|
||||||
| NC score | 0.769563 (rank : 18) | θ value | 2.98157e-11 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15428 | Gene names | HPGD, PGDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
PGDH_MOUSE
|
||||||
| NC score | 0.765576 (rank : 19) | θ value | 1.47974e-10 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8VCC1, Q61106 | Gene names | Hpgd, Pgdh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
DHRS4_MOUSE
|
||||||
| NC score | 0.749386 (rank : 20) | θ value | 4.16044e-13 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99LB2, Q9EQU4 | Gene names | Dhrs4, D14Ucla2 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (mouNRDR). | |||||
|
DHB12_HUMAN
|
||||||
| NC score | 0.748833 (rank : 21) | θ value | 5.62301e-10 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q53GQ0, Q96JU2, Q9Y6G8 | Gene names | HSD17B12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR). | |||||
|
DHRS2_HUMAN
|
||||||
| NC score | 0.748108 (rank : 22) | θ value | 2.36244e-08 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13268, Q9H2R2 | Gene names | DHRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 2 (EC 1.1.-.-) (HEP27 protein) (Protein D). | |||||
|
DHR10_HUMAN
|
||||||
| NC score | 0.746430 (rank : 23) | θ value | 5.08577e-11 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9BPX1, Q9UKU3 | Gene names | DHRS10, SDR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 10 (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase retSDR3). | |||||
|
DHB12_MOUSE
|
||||||
| NC score | 0.740261 (rank : 24) | θ value | 2.13673e-09 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70503, Q3TID1, Q3U7V9, Q542N3, Q8CI39 | Gene names | Hsd17b12, Kik1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 12 (EC 1.1.1.62) (17-beta-HSD 12) (17- beta-hydroxysteroid dehydrogenase 12) (3-ketoacyl-CoA reductase) (EC 1.3.1.-) (KAR) (KIK-I). | |||||
|
CBR2_MOUSE
|
||||||
| NC score | 0.734020 (rank : 25) | θ value | 3.89403e-11 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P08074 | Gene names | Cbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Lung carbonyl reductase [NADPH] (EC 1.1.1.184) (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27). | |||||
|
DHB1_HUMAN
|
||||||
| NC score | 0.729303 (rank : 26) | θ value | 9.59137e-10 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P14061 | Gene names | HSD17B1, E17KSR, EDH17B1, EDHB17 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1) (Placental 17- beta-hydroxysteroid dehydrogenase) (20 alpha-hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHB3_HUMAN
|
||||||
| NC score | 0.725615 (rank : 27) | θ value | 5.62301e-10 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P37058 | Gene names | HSD17B3, EDH17B3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
DCXR_HUMAN
|
||||||
| NC score | 0.716906 (rank : 28) | θ value | 2.13673e-09 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7Z4W1, Q9BTZ3, Q9UHY9 | Gene names | DCXR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase) (Kidney dicarbonyl reductase) (kiDCR) (Carbonyl reductase II) (Sperm surface protein P34H). | |||||
|
DHB1_MOUSE
|
||||||
| NC score | 0.708657 (rank : 29) | θ value | 7.34386e-10 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51656 | Gene names | Hsd17b1, Edh17b1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1). | |||||
|
DCXR_MOUSE
|
||||||
| NC score | 0.703297 (rank : 30) | θ value | 2.36244e-08 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q91X52, Q3U5L5, Q9D129, Q9D8W1 | Gene names | Dcxr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-xylulose reductase (EC 1.1.1.10) (XR) (Dicarbonyl/L-xylulose reductase). | |||||
|
DHB3_MOUSE
|
||||||
| NC score | 0.685568 (rank : 31) | θ value | 1.38499e-08 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
RDH13_MOUSE
|
||||||
| NC score | 0.681284 (rank : 32) | θ value | 9.59137e-10 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8CEE7, Q8CC07 | Gene names | Rdh13 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
DHB8_HUMAN
|
||||||
| NC score | 0.676613 (rank : 33) | θ value | 1.99992e-07 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q92506, Q9UIQ1 | Gene names | HSD17B8, FABGL, HKE6, RING2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
PECR_HUMAN
|
||||||
| NC score | 0.673604 (rank : 34) | θ value | 3.41135e-07 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BY49, Q6IAK9, Q9NRD4, Q9NY60, Q9P1A4 | Gene names | PECR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) (TERP) (HPDHase) (pVI-ARL) (2,4-dienoyl-CoA reductase-related protein) (DCR-RP). | |||||
|
RDH13_HUMAN
|
||||||
| NC score | 0.672749 (rank : 35) | θ value | 2.13673e-09 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8NBN7, Q6UX79, Q96G88 | Gene names | RDH13 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 13 (EC 1.1.1.-). | |||||
|
RDH7_MOUSE
|
||||||
| NC score | 0.660666 (rank : 36) | θ value | 5.62301e-10 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88451 | Gene names | Rdh7, Crad2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinol dehydrogenase 7 (EC 1.1.1.105) (Cis-retinol/androgen dehydrogenase type 2) (CRAD-2) (cis-retinol/3alpha-hydroxysterol short-chain dehydrogenase isozyme 2). | |||||
|
DHRS1_MOUSE
|
||||||
| NC score | 0.660656 (rank : 37) | θ value | 3.19293e-05 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99L04, Q3THW0, Q9D148 | Gene names | Dhrs1, D14ertd484e | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
RDH1_HUMAN
|
||||||
| NC score | 0.655148 (rank : 38) | θ value | 1.06045e-08 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q92781, Q8TAI2 | Gene names | RDH5, RDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 11-cis retinol dehydrogenase (EC 1.1.1.105) (11-cis RDH). | |||||
|
DHRS9_MOUSE
|
||||||
| NC score | 0.650218 (rank : 39) | θ value | 8.11959e-09 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q58NB6, Q8BGC7 | Gene names | Dhrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (Short-chain dehydrogenase/reductase retSDR8). | |||||
|
BDH_MOUSE
|
||||||
| NC score | 0.647446 (rank : 40) | θ value | 1.80886e-08 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q80XN0, Q3UJS9, Q8BK53, Q8R0C8 | Gene names | Bdh1, Bdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
DHRS9_HUMAN
|
||||||
| NC score | 0.637124 (rank : 41) | θ value | 4.76016e-09 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BPW9, Q5RKX1, Q9NRA9, Q9NRB0 | Gene names | DHRS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (RDH-E2) (Short-chain dehydrogenase/reductase retSDR8) (NADP-dependent retinol dehydrogenase/reductase) (RDHL). | |||||
|
PECR_MOUSE
|
||||||
| NC score | 0.635010 (rank : 42) | θ value | 6.43352e-06 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q99MZ7, Q9CX01, Q9JIF4 | Gene names | Pecr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38). | |||||
|
RDH11_HUMAN
|
||||||
| NC score | 0.632746 (rank : 43) | θ value | 3.41135e-07 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8TC12, Q9NRW0, Q9Y391 | Gene names | RDH11, ARSDR1, PSDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (HCV core-binding protein HCBP12). | |||||
|
HCD2_HUMAN
|
||||||
| NC score | 0.631122 (rank : 44) | θ value | 1.99992e-07 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
DHRS1_HUMAN
|
||||||
| NC score | 0.625935 (rank : 45) | θ value | 2.44474e-05 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96LJ7, Q8NDG3, Q96B59, Q96CQ5 | Gene names | DHRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
RDH12_HUMAN
|
||||||
| NC score | 0.622245 (rank : 46) | θ value | 1.133e-10 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96NR8, Q8TAW6 | Gene names | RDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-) (All-trans and 9-cis retinol dehydrogenase). | |||||
|
RDH11_MOUSE
|
||||||
| NC score | 0.620362 (rank : 47) | θ value | 4.45536e-07 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QYF1, Q3UXM9, Q9D0U5 | Gene names | Rdh11, Arsdr1, Mdt1, Psdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 11 (EC 1.1.1.-) (Retinal reductase 1) (RalR1) (Prostate short-chain dehydrogenase/reductase 1) (Androgen-regulated short-chain dehydrogenase/reductase 1) (Short-chain aldehyde dehydrogenase) (SCALD) (Cell line MC/9.IL4-derived protein 1) (M42C60). | |||||
|
DHB4_HUMAN
|
||||||
| NC score | 0.615694 (rank : 48) | θ value | 1.06045e-08 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
RDH12_MOUSE
|
||||||
| NC score | 0.610169 (rank : 49) | θ value | 8.97725e-08 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BYK4, Q91WA5, Q9D1Y4 | Gene names | Rdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 12 (EC 1.1.1.-). | |||||
|
DHB2_MOUSE
|
||||||
| NC score | 0.609721 (rank : 50) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P51658, O08898 | Gene names | Hsd17b2, Edh17b2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (17- beta-hydroxysteroid dehydrogenase 2). | |||||
|
DHI2_MOUSE
|
||||||
| NC score | 0.607012 (rank : 51) | θ value | 4.76016e-09 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51661, Q91WK3 | Gene names | Hsd11b2, Hsd11k | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
DHB4_MOUSE
|
||||||
| NC score | 0.605510 (rank : 52) | θ value | 2.36244e-08 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
BDH_HUMAN
|
||||||
| NC score | 0.605408 (rank : 53) | θ value | 3.41135e-07 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02338, Q96ET1, Q9BRZ4 | Gene names | BDH1, BDH | |||
|
Domain Architecture |

|
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
DHB8_MOUSE
|
||||||
| NC score | 0.588451 (rank : 54) | θ value | 8.40245e-06 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P50171, Q60958, Q60959, Q9Z1W2 | Gene names | Hsd17b8, H2-Ke6, Hke6 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DECR_MOUSE
|
||||||
| NC score | 0.577227 (rank : 55) | θ value | 0.000121331 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9CQ62, Q9DCI7 | Gene names | Decr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
RDH14_HUMAN
|
||||||
| NC score | 0.575203 (rank : 56) | θ value | 1.53129e-07 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HBH5 | Gene names | RDH14, PAN2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
RDH14_MOUSE
|
||||||
| NC score | 0.574118 (rank : 57) | θ value | 1.99992e-07 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9ERI6 | Gene names | Rdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Retinol dehydrogenase 14 (EC 1.1.1.-) (Alcohol dehydrogenase PAN2). | |||||
|
DHRSX_HUMAN
|
||||||
| NC score | 0.569120 (rank : 58) | θ value | 0.000461057 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8N5I4, Q6UWC7, Q8WUS4, Q96GR8, Q9NTF6 | Gene names | DHRSX, CXorf11, DHRS5X | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member on chromosome X precursor (EC 1.1.-.-) (DHRSXY). | |||||
|
DECR_HUMAN
|
||||||
| NC score | 0.564080 (rank : 59) | θ value | 3.77169e-06 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q16698, Q93085 | Gene names | DECR1, DECR | |||
|
Domain Architecture |

|
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
DHB2_HUMAN
|
||||||
| NC score | 0.553406 (rank : 60) | θ value | 5.81887e-07 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P37059 | Gene names | HSD17B2, EDH17B2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 2 (EC 1.1.1.62) (17-beta-HSD 2) (Microsomal 17-beta-hydroxysteroid dehydrogenase) (20 alpha- hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHI2_HUMAN
|
||||||
| NC score | 0.551370 (rank : 61) | θ value | 1.87187e-05 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P80365, Q13194, Q8N439, Q96QN8, Q9UC50, Q9UC51 | Gene names | HSD11B2, HSD11K | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 2 (EC 1.1.1.-) (11-DH2) (11-beta-hydroxysteroid dehydrogenase type 2) (11-beta-HSD2) (NAD- dependent 11-beta-hydroxysteroid dehydrogenase). | |||||
|
HCD2_MOUSE
|
||||||
| NC score | 0.515241 (rank : 62) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
DECR2_HUMAN
|
||||||
| NC score | 0.496773 (rank : 63) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
DHC3_HUMAN
|
||||||
| NC score | 0.491639 (rank : 64) | θ value | 0.00509761 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O75828 | Gene names | CBR3 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 3 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 3). | |||||
|
DECR2_MOUSE
|
||||||
| NC score | 0.478583 (rank : 65) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9WV68 | Gene names | Decr2, Pdcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.462401 (rank : 66) | θ value | 5.81887e-07 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
DHCA_HUMAN
|
||||||
| NC score | 0.453575 (rank : 67) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P16152 | Gene names | CBR1, CBR, CRN | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1) (Prostaglandin-E(2) 9-reductase) (EC 1.1.1.189) (Prostaglandin 9-ketoreductase) (15-hydroxyprostaglandin dehydrogenase [NADP+]) (EC 1.1.1.197). | |||||
|
DHCA_MOUSE
|
||||||
| NC score | 0.439847 (rank : 68) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P48758 | Gene names | Cbr1, Cbr | |||
|
Domain Architecture |

|
|||||
| Description | Carbonyl reductase [NADPH] 1 (EC 1.1.1.184) (NADPH-dependent carbonyl reductase 1). | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.437976 (rank : 69) | θ value | 4.92598e-06 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
DHB7_HUMAN
|
||||||
| NC score | 0.349217 (rank : 70) | θ value | 0.0736092 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P56937, Q7Z4V9, Q8WWS2, Q9UF00 | Gene names | HSD17B7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
DHB7_MOUSE
|
||||||
| NC score | 0.317752 (rank : 71) | θ value | 0.0252991 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O88736, Q8K5C9, Q921L1 | Gene names | Hsd17b7 | |||
|
Domain Architecture |

|
|||||
| Description | 3-keto-steroid reductase (EC 1.1.1.270) (Estradiol 17-beta- dehydrogenase 7) (EC 1.1.1.62) (17-beta-HSD 7) (17-beta-hydroxysteroid dehydrogenase 7). | |||||
|
SPRE_MOUSE
|
||||||
| NC score | 0.244182 (rank : 72) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q64105, Q63996 | Gene names | Spr | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
SPRE_HUMAN
|
||||||
| NC score | 0.209065 (rank : 73) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35270 | Gene names | SPR | |||
|
Domain Architecture |

|
|||||
| Description | Sepiapterin reductase (EC 1.1.1.153) (SPR). | |||||
|
CT079_MOUSE
|
||||||
| NC score | 0.061306 (rank : 74) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DAH1 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf79 homolog. | |||||
|
CT079_HUMAN
|
||||||
| NC score | 0.061289 (rank : 75) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJQ7 | Gene names | C20orf79 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf79. | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.025155 (rank : 76) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.022554 (rank : 77) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
RT4I1_HUMAN
|
||||||
| NC score | 0.016990 (rank : 78) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWV3, Q8N9B3, Q8WZ66, Q9BRA4 | Gene names | RTN4IP1, NIMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4-interacting protein 1, mitochondrial precursor (NOGO- interacting mitochondrial protein). | |||||