Please be patient as the page loads
|
CT079_MOUSE
|
||||||
| SwissProt Accessions | Q9DAH1 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf79 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CT079_MOUSE
|
||||||
| θ value | 3.44927e-84 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DAH1 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf79 homolog. | |||||
|
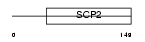
CT079_HUMAN
|
||||||
| θ value | 2.94036e-59 (rank : 2) | NC score | 0.992638 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJQ7 | Gene names | C20orf79 | |||
|
Domain Architecture |
|
|||||
| Description | Uncharacterized protein C20orf79. | |||||
|
DHB4_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 3) | NC score | 0.627707 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
DHB4_MOUSE
|
||||||
| θ value | 6.61148e-27 (rank : 4) | NC score | 0.630223 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
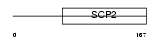
NLTP_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 5) | NC score | 0.615529 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22307, Q15432, Q16622, Q99430 | Gene names | SCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
NLTP_MOUSE
|
||||||
| θ value | 5.43371e-13 (rank : 6) | NC score | 0.607529 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32020, Q9DBM7 | Gene names | Scp2, Scp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
STML1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 7) | NC score | 0.309063 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBI4, O95675, Q6FGL8, Q8WYI7, Q9UMB9, Q9UMC0, Q9Y6H9 | Gene names | STOML1, SLP1, UNC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1) (Stomatin-related protein) (STORP) (EPB72-like 1) (UNC24 homolog). | |||||
|
STML1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 8) | NC score | 0.297056 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CI66, Q8BLA3 | Gene names | Stoml1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1). | |||||
|
MAGL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.011689 (rank : 47) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
CTGE3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.012916 (rank : 46) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX95 | Gene names | CTAGE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 3 (cTAGE-3 protein). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.009535 (rank : 48) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.009443 (rank : 49) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
BDH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 13) | NC score | 0.073660 (rank : 24) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUT1, Q503A0, Q6IA46, Q6UWD3, Q9H8S8, Q9NRX8 | Gene names | BDH2, DHRS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6) (Oxidoreductase UCPA). | |||||
|
BDH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.103583 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8JZV9 | Gene names | Bdh2, Dhrs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6). | |||||
|
BDH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.055157 (rank : 42) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XN0, Q3UJS9, Q8BK53, Q8R0C8 | Gene names | Bdh1, Bdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
CBR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.070491 (rank : 26) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08074 | Gene names | Cbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Lung carbonyl reductase [NADPH] (EC 1.1.1.184) (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27). | |||||
|
DECR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.068695 (rank : 30) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
DECR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.060860 (rank : 35) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQ62, Q9DCI7 | Gene names | Decr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
DHB13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.071617 (rank : 25) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z5P4, Q86W22, Q86W23 | Gene names | HSD17B13, SCDR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9). | |||||
|
DHB13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.061306 (rank : 34) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
DHB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.060034 (rank : 36) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14061 | Gene names | HSD17B1, E17KSR, EDH17B1, EDHB17 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1) (Placental 17- beta-hydroxysteroid dehydrogenase) (20 alpha-hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.070365 (rank : 28) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51656 | Gene names | Hsd17b1, Edh17b1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1). | |||||
|
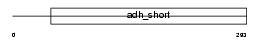
DHB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.077472 (rank : 21) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37058 | Gene names | HSD17B3, EDH17B3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
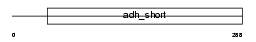
DHB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.059874 (rank : 37) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
DHB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.118254 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92506, Q9UIQ1 | Gene names | HSD17B8, FABGL, HKE6, RING2 | |||
|
Domain Architecture |
|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DHB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.118491 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50171, Q60958, Q60959, Q9Z1W2 | Gene names | Hsd17b8, H2-Ke6, Hke6 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DHI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.055144 (rank : 43) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |
|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
DHR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.067359 (rank : 32) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWP2, Q9BUC7, Q9H674 | Gene names | ames=UNQ836/PRO1774 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHR11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.058177 (rank : 40) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U0B3, Q5SXB3, Q8R249 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHRS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.068697 (rank : 29) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96LJ7, Q8NDG3, Q96B59, Q96CQ5 | Gene names | DHRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DHRS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.079482 (rank : 19) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99L04, Q3THW0, Q9D148 | Gene names | Dhrs1, D14ertd484e | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DHRS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.075456 (rank : 23) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
DHRS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.076133 (rank : 22) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |
|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
DHRS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.079199 (rank : 20) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
DHRS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.081922 (rank : 18) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LB2, Q9EQU4 | Gene names | Dhrs4, D14Ucla2 | |||
|
Domain Architecture |
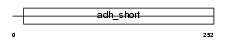
|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (mouNRDR). | |||||
|
DHRS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.056227 (rank : 41) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y394, Q9UKU2 | Gene names | DHRS7, RETSDR4 | |||
|
Domain Architecture |
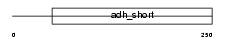
|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4). | |||||
|
DHRS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.058284 (rank : 39) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXR1 | Gene names | Dhrs7, Retsdr4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4). | |||||
|
DHRS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.064036 (rank : 33) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBQ5, Q96HF6, Q9UKU4 | Gene names | DHRS8, HSD17B11, PAN1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11) (Retinal short-chain dehydrogenase/reductase 2) (retSDR2) (Cutaneous T-cell lymphoma- associated antigen HD-CL-03) (CTCL tumor antigen HD-CL-03). | |||||
|
DHRS8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.067831 (rank : 31) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQ06, Q3U2P6, Q8BR33, Q8C7S0 | Gene names | Dhrs8, Hsd17b11, Pan1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11). | |||||
|
FVT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.097590 (rank : 13) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06136 | Gene names | FVT1 | |||
|
Domain Architecture |
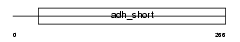
|
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
FVT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.097655 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6GV12 | Gene names | Fvt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
HCD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.083487 (rank : 16) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |
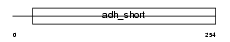
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
HCD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.097358 (rank : 14) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
PECR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.083756 (rank : 15) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY49, Q6IAK9, Q9NRD4, Q9NY60, Q9P1A4 | Gene names | PECR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) (TERP) (HPDHase) (pVI-ARL) (2,4-dienoyl-CoA reductase-related protein) (DCR-RP). | |||||
|
PGDH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.070386 (rank : 27) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15428 | Gene names | HPGD, PGDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
PGDH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.082936 (rank : 17) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCC1, Q61106 | Gene names | Hpgd, Pgdh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
THIK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.050776 (rank : 44) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09110 | Gene names | ACAA1, ACAA, PTHIO | |||
|
Domain Architecture |
|
|||||
| Description | 3-ketoacyl-CoA thiolase, peroxisomal precursor (EC 2.3.1.16) (Beta- ketothiolase) (Acetyl-CoA acyltransferase) (Peroxisomal 3-oxoacyl-CoA thiolase). | |||||
|
THIM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.050511 (rank : 45) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42765, Q9BUT6 | Gene names | ACAA2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) (Beta- ketothiolase) (Acetyl-CoA acyltransferase) (Mitochondrial 3-oxoacyl- CoA thiolase) (T1). | |||||
|
THIM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.058794 (rank : 38) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWT1, Q8JZR8 | Gene names | Acaa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) (Beta- ketothiolase) (Acetyl-CoA acyltransferase) (Mitochondrial 3-oxoacyl- CoA thiolase). | |||||
|
CT079_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.44927e-84 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DAH1 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf79 homolog. | |||||
|
CT079_HUMAN
|
||||||
| NC score | 0.992638 (rank : 2) | θ value | 2.94036e-59 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJQ7 | Gene names | C20orf79 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf79. | |||||
|
DHB4_MOUSE
|
||||||
| NC score | 0.630223 (rank : 3) | θ value | 6.61148e-27 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51660, Q9DBM3 | Gene names | Hsd17b4, Edh17b4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
DHB4_HUMAN
|
||||||
| NC score | 0.627707 (rank : 4) | θ value | 4.57862e-28 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
NLTP_HUMAN
|
||||||
| NC score | 0.615529 (rank : 5) | θ value | 1.09485e-13 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22307, Q15432, Q16622, Q99430 | Gene names | SCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
NLTP_MOUSE
|
||||||
| NC score | 0.607529 (rank : 6) | θ value | 5.43371e-13 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32020, Q9DBM7 | Gene names | Scp2, Scp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
STML1_HUMAN
|
||||||
| NC score | 0.309063 (rank : 7) | θ value | 0.000786445 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBI4, O95675, Q6FGL8, Q8WYI7, Q9UMB9, Q9UMC0, Q9Y6H9 | Gene names | STOML1, SLP1, UNC24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1) (Stomatin-related protein) (STORP) (EPB72-like 1) (UNC24 homolog). | |||||
|
STML1_MOUSE
|
||||||
| NC score | 0.297056 (rank : 8) | θ value | 0.00869519 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CI66, Q8BLA3 | Gene names | Stoml1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stomatin-like protein 1 (SLP-1). | |||||
|
DHB8_MOUSE
|
||||||
| NC score | 0.118491 (rank : 9) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P50171, Q60958, Q60959, Q9Z1W2 | Gene names | Hsd17b8, H2-Ke6, Hke6 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
DHB8_HUMAN
|
||||||
| NC score | 0.118254 (rank : 10) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92506, Q9UIQ1 | Gene names | HSD17B8, FABGL, HKE6, RING2 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 8 (EC 1.1.1.62) (17-beta-HSD 8) (17- beta-hydroxysteroid dehydrogenase 8) (Protein Ke6) (Ke-6). | |||||
|
BDH2_MOUSE
|
||||||
| NC score | 0.103583 (rank : 11) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8JZV9 | Gene names | Bdh2, Dhrs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6). | |||||
|
FVT1_MOUSE
|
||||||
| NC score | 0.097655 (rank : 12) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6GV12 | Gene names | Fvt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
FVT1_HUMAN
|
||||||
| NC score | 0.097590 (rank : 13) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06136 | Gene names | FVT1 | |||
|
Domain Architecture |

|
|||||
| Description | 3-ketodihydrosphingosine reductase precursor (EC 1.1.1.102) (3- dehydrosphinganine reductase) (KDS reductase) (Follicular variant translocation protein 1) (FVT-1). | |||||
|
HCD2_MOUSE
|
||||||
| NC score | 0.097358 (rank : 14) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
PECR_HUMAN
|
||||||
| NC score | 0.083756 (rank : 15) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BY49, Q6IAK9, Q9NRD4, Q9NY60, Q9P1A4 | Gene names | PECR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal trans-2-enoyl-CoA reductase (EC 1.3.1.38) (TERP) (HPDHase) (pVI-ARL) (2,4-dienoyl-CoA reductase-related protein) (DCR-RP). | |||||
|
HCD2_HUMAN
|
||||||
| NC score | 0.083487 (rank : 16) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
PGDH_MOUSE
|
||||||
| NC score | 0.082936 (rank : 17) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCC1, Q61106 | Gene names | Hpgd, Pgdh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
DHRS4_MOUSE
|
||||||
| NC score | 0.081922 (rank : 18) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LB2, Q9EQU4 | Gene names | Dhrs4, D14Ucla2 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (mouNRDR). | |||||
|
DHRS1_MOUSE
|
||||||
| NC score | 0.079482 (rank : 19) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99L04, Q3THW0, Q9D148 | Gene names | Dhrs1, D14ertd484e | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DHRS4_HUMAN
|
||||||
| NC score | 0.079199 (rank : 20) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
DHB3_HUMAN
|
||||||
| NC score | 0.077472 (rank : 21) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P37058 | Gene names | HSD17B3, EDH17B3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
DHRS3_MOUSE
|
||||||
| NC score | 0.076133 (rank : 22) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88876, Q3UAD1, Q91WR0, Q91XC3, Q922A6 | Gene names | Dhrs3, Rsdr1 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1). | |||||
|
DHRS3_HUMAN
|
||||||
| NC score | 0.075456 (rank : 23) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75911, Q6UY38, Q9BUC8 | Gene names | DHRS3 | |||
|
Domain Architecture |

|
|||||
| Description | Short-chain dehydrogenase/reductase 3 (EC 1.1.-.-) (Retinal short- chain dehydrogenase/reductase 1) (retSDR1) (DD83.1). | |||||
|
BDH2_HUMAN
|
||||||
| NC score | 0.073660 (rank : 24) | θ value | θ > 10 (rank : 13) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUT1, Q503A0, Q6IA46, Q6UWD3, Q9H8S8, Q9NRX8 | Gene names | BDH2, DHRS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-hydroxybutyrate dehydrogenase type 2 (EC 1.1.1.30) (R-beta- hydroxybutyrate dehydrogenase) (Dehydrogenase/reductase SDR family member 6) (Oxidoreductase UCPA). | |||||
|
DHB13_HUMAN
|
||||||
| NC score | 0.071617 (rank : 25) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z5P4, Q86W22, Q86W23 | Gene names | HSD17B13, SCDR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9). | |||||
|
CBR2_MOUSE
|
||||||
| NC score | 0.070491 (rank : 26) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08074 | Gene names | Cbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Lung carbonyl reductase [NADPH] (EC 1.1.1.184) (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27). | |||||
|
PGDH_HUMAN
|
||||||
| NC score | 0.070386 (rank : 27) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15428 | Gene names | HPGD, PGDH1 | |||
|
Domain Architecture |

|
|||||
| Description | 15-hydroxyprostaglandin dehydrogenase [NAD+] (EC 1.1.1.141) (PGDH) (Prostaglandin dehydrogenase 1). | |||||
|
DHB1_MOUSE
|
||||||
| NC score | 0.070365 (rank : 28) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51656 | Gene names | Hsd17b1, Edh17b1 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1). | |||||
|
DHRS1_HUMAN
|
||||||
| NC score | 0.068697 (rank : 29) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96LJ7, Q8NDG3, Q96B59, Q96CQ5 | Gene names | DHRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 1 (EC 1.1.-.-). | |||||
|
DECR2_HUMAN
|
||||||
| NC score | 0.068695 (rank : 30) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUI1, Q6ZRS7, Q96ET0 | Gene names | DECR2, PDCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal 2,4-dienoyl-CoA reductase (EC 1.3.1.34) (2,4-dienoyl-CoA reductase 2) (pDCR). | |||||
|
DHRS8_MOUSE
|
||||||
| NC score | 0.067831 (rank : 31) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQ06, Q3U2P6, Q8BR33, Q8C7S0 | Gene names | Dhrs8, Hsd17b11, Pan1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11). | |||||
|
DHR11_HUMAN
|
||||||
| NC score | 0.067359 (rank : 32) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWP2, Q9BUC7, Q9H674 | Gene names | ames=UNQ836/PRO1774 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHRS8_HUMAN
|
||||||
| NC score | 0.064036 (rank : 33) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBQ5, Q96HF6, Q9UKU4 | Gene names | DHRS8, HSD17B11, PAN1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 8 precursor (EC 1.1.1.-) (17-beta-hydroxysteroid dehydrogenase 11) (17-beta-HSD 11) (17-beta- HSD XI) (17betaHSDXI) (17bHSD11) (17betaHSD11) (Retinal short-chain dehydrogenase/reductase 2) (retSDR2) (Cutaneous T-cell lymphoma- associated antigen HD-CL-03) (CTCL tumor antigen HD-CL-03). | |||||
|
DHB13_MOUSE
|
||||||
| NC score | 0.061306 (rank : 34) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCR2, Q8CIU2 | Gene names | Hsd17b13, Scdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 17-beta hydroxysteroid dehydrogenase 13 precursor (EC 1.1.-.-) (Short- chain dehydrogenase/reductase 9) (Alcohol dehydrogenase PAN1B-like). | |||||
|
DECR_MOUSE
|
||||||
| NC score | 0.060860 (rank : 35) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQ62, Q9DCI7 | Gene names | Decr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2,4-dienoyl-CoA reductase, mitochondrial precursor (EC 1.3.1.34) (2,4- dienoyl-CoA reductase [NADPH]) (4-enoyl-CoA reductase [NADPH]). | |||||
|
DHB1_HUMAN
|
||||||
| NC score | 0.060034 (rank : 36) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14061 | Gene names | HSD17B1, E17KSR, EDH17B1, EDHB17 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 1 (EC 1.1.1.62) (17-beta- hydroxysteroid dehydrogenase type 1) (17-beta-HSD 1) (Placental 17- beta-hydroxysteroid dehydrogenase) (20 alpha-hydroxysteroid dehydrogenase) (20-alpha-HSD) (E2DH). | |||||
|
DHB3_MOUSE
|
||||||
| NC score | 0.059874 (rank : 37) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70385 | Gene names | Hsd17b3, Edh17b3 | |||
|
Domain Architecture |

|
|||||
| Description | Estradiol 17-beta-dehydrogenase 3 (EC 1.1.1.62) (17-beta-HSD 3) (Testicular 17-beta-hydroxysteroid dehydrogenase). | |||||
|
THIM_MOUSE
|
||||||
| NC score | 0.058794 (rank : 38) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWT1, Q8JZR8 | Gene names | Acaa2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) (Beta- ketothiolase) (Acetyl-CoA acyltransferase) (Mitochondrial 3-oxoacyl- CoA thiolase). | |||||
|
DHRS7_MOUSE
|
||||||
| NC score | 0.058284 (rank : 39) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXR1 | Gene names | Dhrs7, Retsdr4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4). | |||||
|
DHR11_MOUSE
|
||||||
| NC score | 0.058177 (rank : 40) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U0B3, Q5SXB3, Q8R249 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 11 precursor (EC 1.-.-.-). | |||||
|
DHRS7_HUMAN
|
||||||
| NC score | 0.056227 (rank : 41) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y394, Q9UKU2 | Gene names | DHRS7, RETSDR4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 7 precursor (EC 1.1.-.-) (Retinal short-chain dehydrogenase/reductase 4) (retSDR4). | |||||
|
BDH_MOUSE
|
||||||
| NC score | 0.055157 (rank : 42) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80XN0, Q3UJS9, Q8BK53, Q8R0C8 | Gene names | Bdh1, Bdh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.30) (BDH) (3-hydroxybutyrate dehydrogenase). | |||||
|
DHI1_HUMAN
|
||||||
| NC score | 0.055144 (rank : 43) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28845 | Gene names | HSD11B1, HSD11, HSD11L | |||
|
Domain Architecture |

|
|||||
| Description | Corticosteroid 11-beta-dehydrogenase isozyme 1 (EC 1.1.1.146) (11-DH) (11-beta-hydroxysteroid dehydrogenase 1) (11-beta-HSD1). | |||||
|
THIK_HUMAN
|
||||||
| NC score | 0.050776 (rank : 44) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09110 | Gene names | ACAA1, ACAA, PTHIO | |||
|
Domain Architecture |

|
|||||
| Description | 3-ketoacyl-CoA thiolase, peroxisomal precursor (EC 2.3.1.16) (Beta- ketothiolase) (Acetyl-CoA acyltransferase) (Peroxisomal 3-oxoacyl-CoA thiolase). | |||||
|
THIM_HUMAN
|
||||||
| NC score | 0.050511 (rank : 45) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42765, Q9BUT6 | Gene names | ACAA2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-ketoacyl-CoA thiolase, mitochondrial (EC 2.3.1.16) (Beta- ketothiolase) (Acetyl-CoA acyltransferase) (Mitochondrial 3-oxoacyl- CoA thiolase) (T1). | |||||
|
CTGE3_HUMAN
|
||||||
| NC score | 0.012916 (rank : 46) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IX95 | Gene names | CTAGE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 3 (cTAGE-3 protein). | |||||
|
MAGL2_HUMAN
|
||||||
| NC score | 0.011689 (rank : 47) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UJ55 | Gene names | MAGEL2, NDNL1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGE-like protein 2 (Necdin-like protein 1) (Protein nM15). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.009535 (rank : 48) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.009443 (rank : 49) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||