Please be patient as the page loads
|
APBB2_HUMAN
|
||||||
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
APBB2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987777 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB1_MOUSE
|
||||||
| θ value | 4.25343e-167 (rank : 3) | NC score | 0.950655 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB1_HUMAN
|
||||||
| θ value | 8.86892e-165 (rank : 4) | NC score | 0.952235 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB3_HUMAN
|
||||||
| θ value | 1.48926e-103 (rank : 5) | NC score | 0.937955 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
APBB3_MOUSE
|
||||||
| θ value | 6.25694e-102 (rank : 6) | NC score | 0.937869 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
WWP2_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 7) | NC score | 0.194846 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
WWP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.180191 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 9) | NC score | 0.046005 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 10) | NC score | 0.178827 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 11) | NC score | 0.160470 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 12) | NC score | 0.120677 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 13) | NC score | 0.159203 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 14) | NC score | 0.155784 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.156570 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 16) | NC score | 0.153358 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 17) | NC score | 0.171308 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 18) | NC score | 0.082490 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.104148 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.151277 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.101328 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.113264 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.122400 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.123184 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.080961 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.043093 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GAS7_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.123466 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.010756 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.137668 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
ARH_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.125176 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.057487 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
CI079_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.061844 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.155735 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
ARH_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.124761 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
PIN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.170354 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
PIN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 36) | NC score | 0.188639 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
STON2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 37) | NC score | 0.040644 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.022791 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.019985 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.028667 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
GAS7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.110570 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.024902 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.041568 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.049956 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
PINL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.192009 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.127065 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.021414 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
ZN507_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.003913 (rank : 125) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCN5 | Gene names | ZNF507 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 507. | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.044304 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
K0056_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.024023 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.027450 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.036947 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CAPON_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.105413 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
GNB1L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.017917 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYB4, Q9H2S2, Q9H4M4 | Gene names | GNB1L, GY2, KIAA1645, WDR14 | |||
|
Domain Architecture |
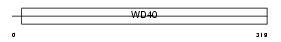
|
|||||
| Description | Guanine nucleotide-binding protein subunit beta-like protein 1 (G protein subunit beta-like protein 1) (WD40 repeat-containing protein deleted in VCFS) (Protein WDVCF) (DGCRK3). | |||||
|
AR13B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.008777 (rank : 119) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
CAPON_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.096002 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.017889 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.012389 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.017110 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.029438 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.014258 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RN123_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.014437 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XPI4, Q5I022, Q6PFW4, Q71RH0, Q8IW18, Q9H0M8, Q9H5L8, Q9H9T2 | Gene names | RNF123, KPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.017241 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
CD44_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.024013 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |
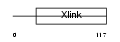
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
ES8L2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.011849 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
HIPK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | -0.000306 (rank : 129) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2X6, Q75MR7, Q8WWI4, Q9H2Y1 | Gene names | HIPK2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (hHIPk2). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.010453 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.013926 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.006756 (rank : 123) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.012860 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
WRN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.009976 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.016726 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ZN451_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.009080 (rank : 116) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.009912 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.024637 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.015855 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.014526 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
K0408_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.017297 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZU52, O43158, Q5TF20, Q7L2M2 | Gene names | KIAA0408 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0408. | |||||
|
PRLR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.008659 (rank : 120) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16471, Q16354, Q9BX87 | Gene names | PRLR | |||
|
Domain Architecture |
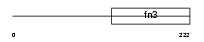
|
|||||
| Description | Prolactin receptor precursor (PRL-R). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.015246 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TAI12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.008909 (rank : 118) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TICN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.014121 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
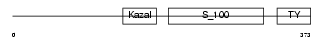
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.003357 (rank : 128) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.003631 (rank : 127) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
ES8L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.010640 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
FA13A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.007736 (rank : 121) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
HTF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.005919 (rank : 124) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.013165 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
MNAB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.009078 (rank : 117) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.003873 (rank : 126) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
PYGO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.009866 (rank : 115) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.011753 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.006885 (rank : 122) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
WWOX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.071287 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.070644 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
ZN346_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.010928 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0B7 | Gene names | Znf346, Jaz, Zfp346 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
BAG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.061917 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.055705 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.061983 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
EDD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.061752 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.056668 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.090397 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.091084 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.062408 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.063513 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.058796 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.059569 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.073186 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
HERC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.074904 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.093857 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.102687 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
K0317_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.095297 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.095206 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.057139 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.060820 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
SAV1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.098206 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
SAV1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.094256 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.115063 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.084564 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.088161 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.087099 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.086804 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.086488 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.069150 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.068695 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.068537 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
WWTR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.057252 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.089533 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.108200 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.987777 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
APBB1_HUMAN
|
||||||
| NC score | 0.952235 (rank : 3) | θ value | 8.86892e-165 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB1_MOUSE
|
||||||
| NC score | 0.950655 (rank : 4) | θ value | 4.25343e-167 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
APBB3_HUMAN
|
||||||
| NC score | 0.937955 (rank : 5) | θ value | 1.48926e-103 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95704, Q9NYX6, Q9NYX7, Q9NYX8 | Gene names | APBB3, FE65L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3 (Fe65-like protein 2) (Fe65L2). | |||||
|
APBB3_MOUSE
|
||||||
| NC score | 0.937869 (rank : 6) | θ value | 6.25694e-102 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8R1C9 | Gene names | Apbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 3. | |||||
|
WWP2_MOUSE
|
||||||
| NC score | 0.194846 (rank : 7) | θ value | 1.17247e-07 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DBH0, Q8BTG4, Q923F6 | Gene names | Wwp2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2). | |||||
|
PINL_HUMAN
|
||||||
| NC score | 0.192009 (rank : 8) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O15428 | Gene names | PIN1L | |||
|
Domain Architecture |

|
|||||
| Description | PIN1-like protein. | |||||
|
PIN1_MOUSE
|
||||||
| NC score | 0.188639 (rank : 9) | θ value | 0.279714 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUR7, Q543B3 | Gene names | Pin1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
WWP1_HUMAN
|
||||||
| NC score | 0.180191 (rank : 10) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H0M0, O00307, Q96BP4 | Gene names | WWP1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1) (Atropin-1-interacting protein 5) (AIP5). | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.178827 (rank : 11) | θ value | 0.00102713 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.171308 (rank : 12) | θ value | 0.0148317 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
PIN1_HUMAN
|
||||||
| NC score | 0.170354 (rank : 13) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13526 | Gene names | PIN1 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (EC 5.2.1.8) (Rotamase Pin1) (PPIase Pin1). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.160470 (rank : 14) | θ value | 0.00509761 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.159203 (rank : 15) | θ value | 0.00509761 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.156570 (rank : 16) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.155784 (rank : 17) | θ value | 0.00665767 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.155735 (rank : 18) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.153358 (rank : 19) | θ value | 0.0148317 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.151277 (rank : 20) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.137668 (rank : 21) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.127065 (rank : 22) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
ARH_HUMAN
|
||||||
| NC score | 0.125176 (rank : 23) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ARH_MOUSE
|
||||||
| NC score | 0.124761 (rank : 24) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
GAS7_HUMAN
|
||||||
| NC score | 0.123466 (rank : 25) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60861, O43144 | Gene names | GAS7, KIAA0394 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.123184 (rank : 26) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.122400 (rank : 27) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.120677 (rank : 28) | θ value | 0.00509761 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.115063 (rank : 29) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.113264 (rank : 30) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
GAS7_MOUSE
|
||||||
| NC score | 0.110570 (rank : 31) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60780, Q9QY25, Q9QY26, Q9QY34 | Gene names | Gas7 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 7 (GAS-7). | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.108200 (rank : 32) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
CAPON_HUMAN
|
||||||
| NC score | 0.105413 (rank : 33) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.104148 (rank : 34) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.102687 (rank : 35) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.101328 (rank : 36) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
SAV1_HUMAN
|
||||||
| NC score | 0.098206 (rank : 37) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H4B6, Q6IA58, Q9H949, Q9HAK9 | Gene names | SAV1, WW45 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (hWW45). | |||||
|
CAPON_MOUSE
|
||||||
| NC score | 0.096002 (rank : 38) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.095297 (rank : 39) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.095206 (rank : 40) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
SAV1_MOUSE
|
||||||
| NC score | 0.094256 (rank : 41) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VEB2, Q9D9N9, Q9ER46 | Gene names | Sav1, Ww45, Wwp3 | |||
|
Domain Architecture |

|
|||||
| Description | Salvador homolog 1 protein (45 kDa WW domain protein) (mWW45). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.093857 (rank : 42) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HECD2_MOUSE
|
||||||
| NC score | 0.091084 (rank : 43) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDU6, Q8CBQ9 | Gene names | Hectd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.090397 (rank : 44) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.089533 (rank : 45) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.088161 (rank : 46) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
UBE3A_MOUSE
|
||||||
| NC score | 0.087099 (rank : 47) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08759, P97482 | Gene names | Ube3a | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (Oncogenic protein- associated protein E6-AP). | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.086804 (rank : 48) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.086488 (rank : 49) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.084564 (rank : 50) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.082490 (rank : 51) | θ value | 0.0193708 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.080961 (rank : 52) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
HERC5_HUMAN
|
||||||
| NC score | 0.074904 (rank : 53) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UII4 | Gene names | HERC5, CEB1, CEBP1 | |||
|
Domain Architecture |

|
|||||
| Description | HECT domain and RCC1-like domain-containing protein 5 (Cyclin-E- binding protein 1). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.073186 (rank : 54) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
WWOX_HUMAN
|
||||||
| NC score | 0.071287 (rank : 55) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZC7, Q5MYT5, Q96KM3, Q96RF2, Q9BTT8, Q9NPC9, Q9NRF4, Q9NRF5, Q9NRF6, Q9NRK1, Q9NZC5 | Gene names | WWOX, FOR, WOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-) (Fragile site FRA16D oxidoreductase). | |||||
|
WWOX_MOUSE
|
||||||
| NC score | 0.070644 (rank : 56) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91WL8, Q8C8J6, Q920Y2, Q9D2B3, Q9D339, Q9JLF5 | Gene names | Wwox, Wox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing oxidoreductase (EC 1.1.1.-). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.069150 (rank : 57) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.068695 (rank : 58) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
WWTR1_HUMAN
|
||||||
| NC score | 0.068537 (rank : 59) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9GZV5, Q8N3P2, Q9Y3W6 | Gene names | WWTR1, TAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
HECD3_MOUSE
|
||||||
| NC score | 0.063513 (rank : 60) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U487, Q3TN76, Q641P3, Q8BQ74, Q8R1L6 | Gene names | Hectd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
HECD3_HUMAN
|
||||||
| NC score | 0.062408 (rank : 61) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T447, Q5T448, Q9H783 | Gene names | HECTD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD3 (HECT domain-containing protein 3). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.061983 (rank : 62) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
BAG3_HUMAN
|
||||||
| NC score | 0.061917 (rank : 63) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95817, Q9NT20, Q9P120 | Gene names | BAG3, BIS | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis) (Docking protein CAIR-1). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.061844 (rank : 64) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
EDD1_MOUSE
|
||||||
| NC score | 0.061752 (rank : 65) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80TP3, Q698K9, Q6PEQ8, Q6PFQ9, Q80VL4, Q810V6, Q9CXE9 | Gene names | Edd1, Edd, Kiaa0896 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.060820 (rank : 66) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
HERC2_MOUSE
|
||||||
| NC score | 0.059569 (rank : 67) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4U2R1, O88473, Q3TRJ8, Q3TS47, Q3TST2, Q3UFQ6, Q3URH7, Q5DU32, Q7TPR5, Q80VV7, Q9QYT1, Q9Z168, Q9Z171 | Gene names | Herc2, jdf2, Kiaa0393, rjs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
HERC2_HUMAN
|
||||||
| NC score | 0.058796 (rank : 68) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95714, Q86SV7, Q86SV8, Q86SV9, Q86YY3, Q86YY4, Q86YY5, Q86YY6, Q86YY7, Q86YY8, Q86YY9, Q86YZ0, Q86YZ1 | Gene names | HERC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain-containing protein 2. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.057487 (rank : 69) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
WWTR1_MOUSE
|
||||||
| NC score | 0.057252 (rank : 70) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9EPK5, Q99KI4 | Gene names | Wwtr1, Taz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing transcription regulator protein 1 (Transcriptional coactivator with PDZ-binding motif). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.057139 (rank : 71) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.056668 (rank : 72) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.055705 (rank : 73) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.049956 (rank : 74) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.046005 (rank : 75) | θ value | 0.00102713 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.044304 (rank : 76) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.043093 (rank : 77) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.041568 (rank : 78) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.040644 (rank : 79) | θ value | 0.279714 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.036947 (rank : 80) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.029438 (rank : 81) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.028667 (rank : 82) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.027450 (rank : 83) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.024902 (rank : 84) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.024637 (rank : 85) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.024023 (rank : 86) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
CD44_HUMAN
|
||||||
| NC score | 0.024013 (rank : 87) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.022791 (rank : 88) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.021414 (rank : 89) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.019985 (rank : 90) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GNB1L_HUMAN
|
||||||
| NC score | 0.017917 (rank : 91) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYB4, Q9H2S2, Q9H4M4 | Gene names | GNB1L, GY2, KIAA1645, WDR14 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein subunit beta-like protein 1 (G protein subunit beta-like protein 1) (WD40 repeat-containing protein deleted in VCFS) (Protein WDVCF) (DGCRK3). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.017889 (rank : 92) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
K0408_HUMAN
|
||||||
| NC score | 0.017297 (rank : 93) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZU52, O43158, Q5TF20, Q7L2M2 | Gene names | KIAA0408 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0408. | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.017241 (rank : 94) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.017110 (rank : 95) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.016726 (rank : 96) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.015855 (rank : 97) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.015246 (rank : 98) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.014526 (rank : 99) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
RN123_HUMAN
|
||||||
| NC score | 0.014437 (rank : 100) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XPI4, Q5I022, Q6PFW4, Q71RH0, Q8IW18, Q9H0M8, Q9H5L8, Q9H9T2 | Gene names | RNF123, KPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase subunit KPC1 (EC 6.3.2.-) (Kip1 ubiquitination-promoting complex protein 1) (RING finger protein 123). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.014258 (rank : 101) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.014121 (rank : 102) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.013926 (rank : 103) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.013165 (rank : 104) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.012860 (rank : 105) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.012389 (rank : 106) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
ES8L2_MOUSE
|
||||||
| NC score | 0.011849 (rank : 107) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99K30, Q91VT7 | Gene names | Eps8l2, Eps8r2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.011753 (rank : 108) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
ZN346_MOUSE
|
||||||
| NC score | 0.010928 (rank : 109) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0B7 | Gene names | Znf346, Jaz, Zfp346 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 346 (Just another zinc finger protein). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.010756 (rank : 110) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
ES8L2_HUMAN
|
||||||
| NC score | 0.010640 (rank : 111) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H6S3, Q53GM8, Q8WYW7, Q96K06, Q9H6K9 | Gene names | EPS8L2, EPS8R2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 2 (Epidermal growth factor receptor pathway substrate 8-related protein 2) (EPS8-like protein 2). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.010453 (rank : 112) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
WRN_MOUSE
|
||||||
| NC score | 0.009976 (rank : 113) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O09053, O09050, Q9JKD4, Q9Z241, Q9Z242 | Gene names | Wrn | |||
|
Domain Architecture |

|
|||||
| Description | Werner syndrome ATP-dependent helicase homolog (EC 3.6.1.-). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.009912 (rank : 114) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PYGO1_HUMAN
|
||||||
| NC score | 0.009866 (rank : 115) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
ZN451_MOUSE
|
||||||
| NC score | 0.009080 (rank : 116) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0P7 | Gene names | Znf451, Zfp451 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 451. | |||||
|
MNAB_MOUSE
|
||||||
| NC score | 0.009078 (rank : 117) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P0C090 | Gene names | Mnab | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated nucleic acid-binding protein. | |||||
|
TAI12_HUMAN
|
||||||
| NC score | 0.008909 (rank : 118) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
AR13B_MOUSE
|
||||||
| NC score | 0.008777 (rank : 119) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
PRLR_HUMAN
|
||||||
| NC score | 0.008659 (rank : 120) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P16471, Q16354, Q9BX87 | Gene names | PRLR | |||
|
Domain Architecture |

|
|||||
| Description | Prolactin receptor precursor (PRL-R). | |||||
|
FA13A_HUMAN
|
||||||
| NC score | 0.007736 (rank : 121) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94988, Q8NBA3 | Gene names | FAM13A1, KIAA0914 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13A1. | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.006885 (rank : 122) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.006756 (rank : 123) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.005919 (rank : 124) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
ZN507_HUMAN
|
||||||
| NC score | 0.003913 (rank : 125) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCN5 | Gene names | ZNF507 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 507. | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.003873 (rank : 126) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.003631 (rank : 127) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.003357 (rank : 128) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
HIPK2_HUMAN
|
||||||
| NC score | -0.000306 (rank : 129) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2X6, Q75MR7, Q8WWI4, Q9H2Y1 | Gene names | HIPK2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (hHIPk2). | |||||