Please be patient as the page loads
|
ISK5_HUMAN
|
||||||
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ISK5_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 2) | NC score | 0.216150 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ISK6_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 3) | NC score | 0.540270 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
IPK1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 4) | NC score | 0.554793 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
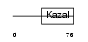
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 5) | NC score | 0.528353 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
FST_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 6) | NC score | 0.363140 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
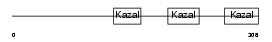
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 7) | NC score | 0.527975 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
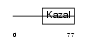
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
ISK4_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 8) | NC score | 0.486165 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |
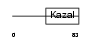
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 9) | NC score | 0.300037 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 10) | NC score | 0.297296 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
IPK2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 11) | NC score | 0.455618 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
ISK4_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.481143 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |
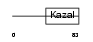
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
ISK7_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 13) | NC score | 0.455036 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |
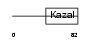
|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.338866 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
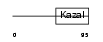
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 15) | NC score | 0.336552 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
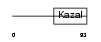
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 16) | NC score | 0.060269 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FST_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.344107 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
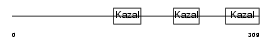
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
IPK2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.459405 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |
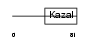
|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
TICN3_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.230326 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
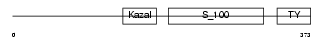
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 20) | NC score | 0.231905 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
ISK7_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 21) | NC score | 0.410744 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 22) | NC score | 0.036336 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
RECK_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.150503 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
RECK_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.142744 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.201467 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
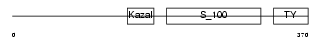
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.093647 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
TICN2_MOUSE
|
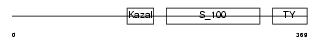
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.202846 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
ADA32_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.022761 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K410, Q6P901, Q8BJ80 | Gene names | Adam32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 32 precursor (A disintegrin and metalloproteinase domain 32). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.025252 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.178717 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.261070 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.125537 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
FSTL5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.124248 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.119971 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
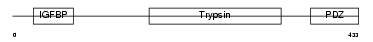
HTRA3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.094151 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
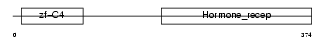
NR2E1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.031060 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64104 | Gene names | Nr2e1, Tll, Tlx | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (mTll). | |||||
|
PARP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.039680 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
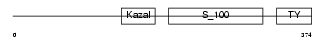
TICN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.191933 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ZN394_HUMAN
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.007349 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q53GI3, Q6P5X9, Q8TB27, Q9HA37 | Gene names | ZNF394 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 394. | |||||
|
ZN668_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.007694 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96K58, Q59EV1, Q8N669, Q9H8L4 | Gene names | ZNF668 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 668. | |||||
|
ZN668_MOUSE
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.007713 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2R5, Q3TEM2, Q3U7Y5, Q8C8B7 | Gene names | Znf668, Zfp668 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 668. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.035655 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NR2E1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.030792 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y466 | Gene names | NR2E1, TLX | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (hTll). | |||||
|
PTN12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.016693 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
TNR9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.038256 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
FA76B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.052033 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XP8, Q3UMA6, Q80YR1, Q8CI69 | Gene names | Fam76b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
ITB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.028358 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.180001 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.015850 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.150113 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.020987 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
FA76B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.048547 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HYJ3, Q6PIU3, Q8TC53 | Gene names | FAM76B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.121098 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.250376 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.121953 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
RARA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.025011 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11416, P22603 | Gene names | Rara, Nr1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.024911 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22605, P11417, P22604 | Gene names | Rarb, Nr1b2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor beta (RAR-beta). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.035075 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
SO5A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.038347 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2Y9 | Gene names | SLCO5A1, SLC21A15 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 5A1 (Solute carrier family 21 member 15) (Organic anion transporter polypeptide- related protein 4) (OATP-RP4) (OATPRP4). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.077687 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
RARB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.024483 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10826, P12891, Q00989, Q15298, Q9UN48 | Gene names | RARB, HAP, NR1B2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor beta (RAR-beta) (RAR-epsilon) (HBV-activated protein). | |||||
|
ITB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.025512 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.019414 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.017635 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.027664 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
SO4A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.028806 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K078, Q8BZT4 | Gene names | Slco4a1, Oatpe, Slc21a12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E). | |||||
|
SPRC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.159954 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.157906 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.084972 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.007108 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.017057 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ZN260_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.004178 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62513, Q3U170, Q8C2Q6, Q9R2B4 | Gene names | Znf260, Zfp260 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 260 (Zfp-260). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.014642 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
IBP7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.080837 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
LIPR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.011630 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5BKQ4, O70478, Q6NV82 | Gene names | Pnliprp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreatic lipase-related protein 1 precursor (EC 3.1.1.3). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.018241 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.018257 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
RARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.022664 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.011343 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
ZN711_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.005604 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y462, Q6NX42, Q9Y4J6 | Gene names | ZNF711, CMPX1, ZNF6 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 711 (Zinc finger protein 6). | |||||
|
1B53_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.000837 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30491, O78053, O78138, Q30198, Q9BD43, Q9GJF0, Q9MY42, Q9TP36, Q9TQH8 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |
|
|||||
| Description | HLA class I histocompatibility antigen, B-53 alpha chain precursor (MHC class I antigen B*53) (Bw-53). | |||||
|
ADA2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | -0.000649 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 881 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18089, Q4TUH9, Q53RF2, Q9BZK0 | Gene names | ADRA2B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor) (Alpha-2 adrenergic receptor subtype C2). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.013165 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
NOL7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.023317 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7Z3, Q9CY79, Q9JJE2 | Gene names | Nol7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 7. | |||||
|
PDE1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.008708 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
SO1A6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.019890 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99J94, Q3UQC6, Q9CUP7 | Gene names | Slco1a6, Oatp5, Slc21a13 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1A6 (Solute carrier family 21 member 13) (Kidney-specific organic anion- transporting polypeptide 5) (OATP-5). | |||||
|
ZFP38_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.004227 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07231 | Gene names | Znf38, Zfp-38, Zfp38, Zipro1 | |||
|
Domain Architecture |
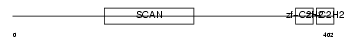
|
|||||
| Description | Zinc finger protein 38 (Zfp-38) (CtFIN51) (Transcription factor RU49). | |||||
|
ZFY1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.005177 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
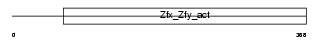
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ZRF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.014866 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.075941 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
IBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.072457 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
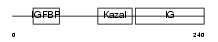
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.089658 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.082972 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.103862 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
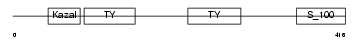
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.107114 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
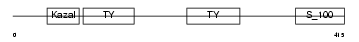
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.104052 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
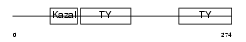
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.554793 (rank : 2) | θ value | 3.77169e-06 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
ISK6_MOUSE
|
||||||
| NC score | 0.540270 (rank : 3) | θ value | 2.61198e-07 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.528353 (rank : 4) | θ value | 3.77169e-06 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.527975 (rank : 5) | θ value | 8.40245e-06 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
ISK4_MOUSE
|
||||||
| NC score | 0.486165 (rank : 6) | θ value | 5.44631e-05 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
ISK4_HUMAN
|
||||||
| NC score | 0.481143 (rank : 7) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
IPK2_HUMAN
|
||||||
| NC score | 0.459405 (rank : 8) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
IPK2_MOUSE
|
||||||
| NC score | 0.455618 (rank : 9) | θ value | 0.00035302 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
ISK7_HUMAN
|
||||||
| NC score | 0.455036 (rank : 10) | θ value | 0.000602161 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
ISK7_MOUSE
|
||||||
| NC score | 0.410744 (rank : 11) | θ value | 0.00869519 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.363140 (rank : 12) | θ value | 8.40245e-06 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.344107 (rank : 13) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.338866 (rank : 14) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.336552 (rank : 15) | θ value | 0.00102713 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.300037 (rank : 16) | θ value | 0.00020696 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.297296 (rank : 17) | θ value | 0.000270298 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.261070 (rank : 18) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.250376 (rank : 19) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.231905 (rank : 20) | θ value | 0.00665767 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.230326 (rank : 21) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.216150 (rank : 22) | θ value | 4.16044e-13 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.202846 (rank : 23) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.201467 (rank : 24) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.191933 (rank : 25) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.180001 (rank : 26) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.178717 (rank : 27) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.159954 (rank : 28) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.157906 (rank : 29) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.150503 (rank : 30) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.150113 (rank : 31) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.142744 (rank : 32) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.125537 (rank : 33) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
FSTL5_HUMAN
|
||||||
| NC score | 0.124248 (rank : 34) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.121953 (rank : 35) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.121098 (rank : 36) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.119971 (rank : 37) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.107114 (rank : 38) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.104052 (rank : 39) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.103862 (rank : 40) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.094151 (rank : 41) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.093647 (rank : 42) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.089658 (rank : 43) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.084972 (rank : 44) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.082972 (rank : 45) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.080837 (rank : 46) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.077687 (rank : 47) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.075941 (rank : 48) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.072457 (rank : 49) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.060269 (rank : 50) | θ value | 0.00102713 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
FA76B_MOUSE
|
||||||
| NC score | 0.052033 (rank : 51) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XP8, Q3UMA6, Q80YR1, Q8CI69 | Gene names | Fam76b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
FA76B_HUMAN
|
||||||
| NC score | 0.048547 (rank : 52) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5HYJ3, Q6PIU3, Q8TC53 | Gene names | FAM76B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
PARP1_MOUSE
|
||||||
| NC score | 0.039680 (rank : 53) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11103, Q9JLX4, Q9QVQ3 | Gene names | Parp1, Adprp, Adprt, Adprt1 | |||
|
Domain Architecture |

|
|||||
| Description | Poly [ADP-ribose] polymerase 1 (EC 2.4.2.30) (PARP-1) (ADPRT) (NAD(+) ADP-ribosyltransferase 1) (Poly[ADP-ribose] synthetase 1) (msPARP). | |||||
|
SO5A1_HUMAN
|
||||||
| NC score | 0.038347 (rank : 54) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H2Y9 | Gene names | SLCO5A1, SLC21A15 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 5A1 (Solute carrier family 21 member 15) (Organic anion transporter polypeptide- related protein 4) (OATP-RP4) (OATPRP4). | |||||
|
TNR9_HUMAN
|
||||||
| NC score | 0.038256 (rank : 55) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.036336 (rank : 56) | θ value | 0.0113563 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.035655 (rank : 57) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.035075 (rank : 58) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
NR2E1_MOUSE
|
||||||
| NC score | 0.031060 (rank : 59) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64104 | Gene names | Nr2e1, Tll, Tlx | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (mTll). | |||||
|
NR2E1_HUMAN
|
||||||
| NC score | 0.030792 (rank : 60) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y466 | Gene names | NR2E1, TLX | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (hTll). | |||||
|
SO4A1_MOUSE
|
||||||
| NC score | 0.028806 (rank : 61) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K078, Q8BZT4 | Gene names | Slco4a1, Oatpe, Slc21a12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E). | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.028358 (rank : 62) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.027664 (rank : 63) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
ITB1_MOUSE
|
||||||
| NC score | 0.025512 (rank : 64) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.025252 (rank : 65) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
RARA_MOUSE
|
||||||
| NC score | 0.025011 (rank : 66) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11416, P22603 | Gene names | Rara, Nr1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARB_MOUSE
|
||||||
| NC score | 0.024911 (rank : 67) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22605, P11417, P22604 | Gene names | Rarb, Nr1b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor beta (RAR-beta). | |||||
|
RARB_HUMAN
|
||||||
| NC score | 0.024483 (rank : 68) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10826, P12891, Q00989, Q15298, Q9UN48 | Gene names | RARB, HAP, NR1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor beta (RAR-beta) (RAR-epsilon) (HBV-activated protein). | |||||
|
NOL7_MOUSE
|
||||||
| NC score | 0.023317 (rank : 69) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7Z3, Q9CY79, Q9JJE2 | Gene names | Nol7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 7. | |||||
|
ADA32_MOUSE
|
||||||
| NC score | 0.022761 (rank : 70) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K410, Q6P901, Q8BJ80 | Gene names | Adam32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 32 precursor (A disintegrin and metalloproteinase domain 32). | |||||
|
RARA_HUMAN
|
||||||
| NC score | 0.022664 (rank : 71) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.020987 (rank : 72) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
SO1A6_MOUSE
|
||||||
| NC score | 0.019890 (rank : 73) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99J94, Q3UQC6, Q9CUP7 | Gene names | Slco1a6, Oatp5, Slc21a13 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1A6 (Solute carrier family 21 member 13) (Kidney-specific organic anion- transporting polypeptide 5) (OATP-5). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.019414 (rank : 74) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.018257 (rank : 75) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.018241 (rank : 76) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.017635 (rank : 77) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.017057 (rank : 78) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
PTN12_HUMAN
|
||||||
| NC score | 0.016693 (rank : 79) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05209, Q16130, Q59FD6, Q75MN8, Q86XU4 | Gene names | PTPN12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase G1) (PTPG1). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.015850 (rank : 80) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
ZRF1_MOUSE
|
||||||
| NC score | 0.014866 (rank : 81) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54103 | Gene names | Zrf1, Dnajc2 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1. | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.014642 (rank : 82) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.013165 (rank : 83) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
LIPR1_MOUSE
|
||||||
| NC score | 0.011630 (rank : 84) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5BKQ4, O70478, Q6NV82 | Gene names | Pnliprp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreatic lipase-related protein 1 precursor (EC 3.1.1.3). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.011343 (rank : 85) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
PDE1C_MOUSE
|
||||||
| NC score | 0.008708 (rank : 86) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64338, Q62045, Q8BSV6 | Gene names | Pde1c | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1C (EC 3.1.4.17) (Cam-PDE 1C). | |||||
|
ZN668_MOUSE
|
||||||
| NC score | 0.007713 (rank : 87) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K2R5, Q3TEM2, Q3U7Y5, Q8C8B7 | Gene names | Znf668, Zfp668 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 668. | |||||
|
ZN668_HUMAN
|
||||||
| NC score | 0.007694 (rank : 88) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96K58, Q59EV1, Q8N669, Q9H8L4 | Gene names | ZNF668 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 668. | |||||
|
ZN394_HUMAN
|
||||||
| NC score | 0.007349 (rank : 89) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q53GI3, Q6P5X9, Q8TB27, Q9HA37 | Gene names | ZNF394 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 394. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.007108 (rank : 90) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
ZN711_HUMAN
|
||||||
| NC score | 0.005604 (rank : 91) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y462, Q6NX42, Q9Y4J6 | Gene names | ZNF711, CMPX1, ZNF6 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 711 (Zinc finger protein 6). | |||||
|
ZFY1_MOUSE
|
||||||
| NC score | 0.005177 (rank : 92) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10925 | Gene names | Zfy1, Zfy-1 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Y-chromosomal protein 1. | |||||
|
ZFP38_MOUSE
|
||||||
| NC score | 0.004227 (rank : 93) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07231 | Gene names | Znf38, Zfp-38, Zfp38, Zipro1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 38 (Zfp-38) (CtFIN51) (Transcription factor RU49). | |||||
|
ZN260_MOUSE
|
||||||
| NC score | 0.004178 (rank : 94) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62513, Q3U170, Q8C2Q6, Q9R2B4 | Gene names | Znf260, Zfp260 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 260 (Zfp-260). | |||||
|
1B53_HUMAN
|
||||||
| NC score | 0.000837 (rank : 95) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30491, O78053, O78138, Q30198, Q9BD43, Q9GJF0, Q9MY42, Q9TP36, Q9TQH8 | Gene names | HLA-B, HLAB | |||
|
Domain Architecture |

|
|||||
| Description | HLA class I histocompatibility antigen, B-53 alpha chain precursor (MHC class I antigen B*53) (Bw-53). | |||||
|
ADA2B_HUMAN
|
||||||
| NC score | -0.000649 (rank : 96) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 881 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18089, Q4TUH9, Q53RF2, Q9BZK0 | Gene names | ADRA2B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2B adrenergic receptor (Alpha-2B adrenoceptor) (Alpha-2B adrenoreceptor) (Alpha-2 adrenergic receptor subtype C2). | |||||