Please be patient as the page loads
|
FSTL1_HUMAN
|
||||||
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FSTL1_HUMAN
|
||||||
| θ value | 3.04821e-165 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | 1.08415e-162 (rank : 2) | NC score | 0.998705 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 3) | NC score | 0.429062 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
FSTL5_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 4) | NC score | 0.416446 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 5) | NC score | 0.412613 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 6) | NC score | 0.426169 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
FST_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 7) | NC score | 0.609516 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 8) | NC score | 0.608423 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 9) | NC score | 0.277090 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 10) | NC score | 0.545881 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 11) | NC score | 0.555192 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 12) | NC score | 0.350714 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 13) | NC score | 0.398946 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 14) | NC score | 0.419234 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 15) | NC score | 0.418745 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
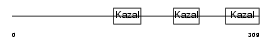
TICN3_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 16) | NC score | 0.380605 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
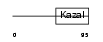
Domain Architecture |
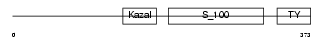
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 17) | NC score | 0.141588 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
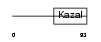
|
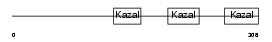
TICN3_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 18) | NC score | 0.383245 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
SPRC_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 19) | NC score | 0.382279 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
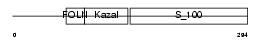
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 20) | NC score | 0.368091 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
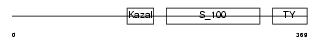
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 21) | NC score | 0.357790 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
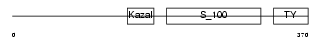
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
SPRC_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 22) | NC score | 0.381717 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
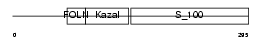
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 23) | NC score | 0.338866 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 24) | NC score | 0.104971 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.203604 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.314686 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
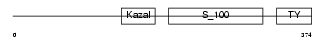
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.058191 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 28) | NC score | 0.274018 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
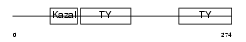
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.301694 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
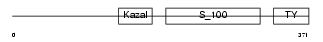
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.378631 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK7_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 31) | NC score | 0.305917 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
RECK_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 32) | NC score | 0.200521 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
VWF_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 33) | NC score | 0.128686 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.270033 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 35) | NC score | 0.129987 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
IBP7_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.192073 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.121258 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
RECK_MOUSE
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.188785 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
TINAG_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.070387 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.040297 (rank : 93) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
ISK4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.318952 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.201180 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.039379 (rank : 94) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.272662 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
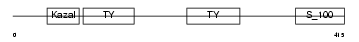
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
IPK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 45) | NC score | 0.274685 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |
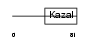
|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
ISK7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 46) | NC score | 0.300699 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |
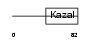
|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
NCS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 47) | NC score | 0.032208 (rank : 98) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
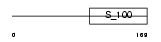
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.032208 (rank : 99) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
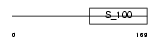
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
ISK6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.312504 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
IBP7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.181034 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
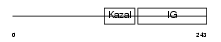
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
ISK4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.267968 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |
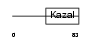
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.108430 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.037396 (rank : 96) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.259282 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
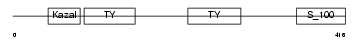
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
CFAI_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.029309 (rank : 102) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.083040 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.038305 (rank : 95) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
ATS20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.014628 (rank : 107) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.031055 (rank : 100) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
CD244_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.028764 (rank : 103) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07763, O88654, Q9JIE0 | Gene names | Cd244, 2b4, Nmrk | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell receptor 2B4 precursor (NKR2B4) (NK cell type I receptor protein 2B4) (CD244 antigen) (Non MHC restricted killing associated). | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.092637 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
IPK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.191508 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
SO5A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.030971 (rank : 101) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H2Y9 | Gene names | SLCO5A1, SLC21A15 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 5A1 (Solute carrier family 21 member 15) (Organic anion transporter polypeptide- related protein 4) (OATP-RP4) (OATPRP4). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.033452 (rank : 97) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
SO1B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.024382 (rank : 104) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6L6, Q9NQ37, Q9UBF3, Q9UH89 | Gene names | SLCO1B1, LST1, OATP2, OATPC, SLC21A6 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1B1 (Solute carrier family 21 member 6) (Sodium-independent organic anion- transporting polypeptide 2) (OATP 2) (Liver-specific organic anion transporter 1) (LST-1) (OATP-C). | |||||
|
SO1B3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.022881 (rank : 105) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPD5 | Gene names | SLCO1B3, LST2, SLC21A8 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1B3 (Solute carrier family 21 member 8) (Organic anion transporter 8) (Organic anion-transporting polypeptide 8) (OATP8) (Liver-specific organic anion transporter 2) (LST-2). | |||||
|
SORL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.022094 (rank : 106) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.082409 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BMPER_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.073222 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.070869 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
CI094_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.057414 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q496M8, Q496M6, Q5VZT8, Q8NAI9 | Gene names | C9orf94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf94 precursor. | |||||
|
HG2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.084937 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
HG2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.088396 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.067940 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.064623 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.056683 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.053630 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051893 (rank : 88) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.050474 (rank : 91) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.068164 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.058961 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.056813 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.055552 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.063174 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.216043 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
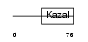
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.181587 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
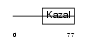
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
JAM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.062806 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57087, Q6UXG6, Q6YNC1 | Gene names | JAM2, C21orf43, VEJAM | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule B precursor (JAM-B) (Junctional adhesion molecule 2) (Vascular endothelial junction-associated molecule) (VE- JAM) (CD322 antigen). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.052271 (rank : 87) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.054253 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR104_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.073896 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.051132 (rank : 90) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.060397 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR109_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.059643 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.056492 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.052937 (rank : 85) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.059070 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
MOX2R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052514 (rank : 86) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES57 | Gene names | Cd200r1, Mox2r, Ox2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell surface glycoprotein OX2 receptor precursor (CD200 cell surface glycoprotein receptor). | |||||
|
NRG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.054325 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
NRG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.054737 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56974 | Gene names | Nrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Divergent of neuregulin 1) (DON-1)]. | |||||
|
PUNC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.051235 (rank : 89) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVU1, O95215 | Gene names | PUNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
PUNC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050418 (rank : 92) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
TACD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054045 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
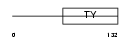
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
TECTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.072176 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.066879 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TMIGD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.069911 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXZ0, Q6ZMC6 | Gene names | TMIGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and immunoglobulin domain-containing protein precursor. | |||||
|
TMIGD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.063949 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D7L8, Q8R202 | Gene names | Tmigd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and immunoglobulin domain-containing protein precursor. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.070962 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.04821e-165 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.998705 (rank : 2) | θ value | 1.08415e-162 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.609516 (rank : 3) | θ value | 5.43371e-13 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.608423 (rank : 4) | θ value | 7.09661e-13 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.555192 (rank : 5) | θ value | 1.99992e-07 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.545881 (rank : 6) | θ value | 1.53129e-07 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.429062 (rank : 7) | θ value | 4.29542e-18 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.426169 (rank : 8) | θ value | 3.76295e-14 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.419234 (rank : 9) | θ value | 1.09739e-05 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.418745 (rank : 10) | θ value | 1.43324e-05 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
FSTL5_HUMAN
|
||||||
| NC score | 0.416446 (rank : 11) | θ value | 7.32683e-18 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.412613 (rank : 12) | θ value | 8.10077e-17 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.398946 (rank : 13) | θ value | 8.40245e-06 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.383245 (rank : 14) | θ value | 4.1701e-05 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.382279 (rank : 15) | θ value | 0.000270298 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.381717 (rank : 16) | θ value | 0.000786445 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.380605 (rank : 17) | θ value | 1.87187e-05 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.378631 (rank : 18) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.368091 (rank : 19) | θ value | 0.000270298 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.357790 (rank : 20) | θ value | 0.000461057 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.350714 (rank : 21) | θ value | 4.92598e-06 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.338866 (rank : 22) | θ value | 0.00102713 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
ISK4_HUMAN
|
||||||
| NC score | 0.318952 (rank : 23) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.314686 (rank : 24) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ISK6_MOUSE
|
||||||
| NC score | 0.312504 (rank : 25) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK7_MOUSE
|
||||||
| NC score | 0.305917 (rank : 26) | θ value | 0.0252991 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.301694 (rank : 27) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ISK7_HUMAN
|
||||||
| NC score | 0.300699 (rank : 28) | θ value | 0.47712 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.277090 (rank : 29) | θ value | 2.13673e-09 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
IPK2_HUMAN
|
||||||
| NC score | 0.274685 (rank : 30) | θ value | 0.47712 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.274018 (rank : 31) | θ value | 0.0113563 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.272662 (rank : 32) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.270033 (rank : 33) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
ISK4_MOUSE
|
||||||
| NC score | 0.267968 (rank : 34) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.259282 (rank : 35) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.216043 (rank : 36) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.203604 (rank : 37) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.201180 (rank : 38) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.200521 (rank : 39) | θ value | 0.0330416 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.192073 (rank : 40) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
IPK2_MOUSE
|
||||||
| NC score | 0.191508 (rank : 41) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.188785 (rank : 42) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.181587 (rank : 43) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.181034 (rank : 44) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.141588 (rank : 45) | θ value | 2.44474e-05 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.129987 (rank : 46) | θ value | 0.0431538 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.128686 (rank : 47) | θ value | 0.0330416 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.121258 (rank : 48) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.108430 (rank : 49) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.104971 (rank : 50) | θ value | 0.00102713 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.092637 (rank : 51) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.088396 (rank : 52) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
HG2A_HUMAN
|
||||||
| NC score | 0.084937 (rank : 53) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.083040 (rank : 54) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.082409 (rank : 55) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.073896 (rank : 56) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.073222 (rank : 57) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.072176 (rank : 58) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.070962 (rank : 59) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.070869 (rank : 60) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.070387 (rank : 61) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
TMIGD_HUMAN
|
||||||
| NC score | 0.069911 (rank : 62) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6UXZ0, Q6ZMC6 | Gene names | TMIGD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and immunoglobulin domain-containing protein precursor. | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.068164 (rank : 63) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.067940 (rank : 64) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.066879 (rank : 65) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.064623 (rank : 66) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
TMIGD_MOUSE
|
||||||
| NC score | 0.063949 (rank : 67) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D7L8, Q8R202 | Gene names | Tmigd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and immunoglobulin domain-containing protein precursor. | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.063174 (rank : 68) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
JAM2_HUMAN
|
||||||
| NC score | 0.062806 (rank : 69) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P57087, Q6UXG6, Q6YNC1 | Gene names | JAM2, C21orf43, VEJAM | |||
|
Domain Architecture |

|
|||||
| Description | Junctional adhesion molecule B precursor (JAM-B) (Junctional adhesion molecule 2) (Vascular endothelial junction-associated molecule) (VE- JAM) (CD322 antigen). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.060397 (rank : 70) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.059643 (rank : 71) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.059070 (rank : 72) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.058961 (rank : 73) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.058191 (rank : 74) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CI094_HUMAN
|
||||||
| NC score | 0.057414 (rank : 75) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q496M8, Q496M6, Q5VZT8, Q8NAI9 | Gene names | C9orf94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf94 precursor. | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.056813 (rank : 76) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.056683 (rank : 77) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.056492 (rank : 78) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.055552 (rank : 79) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
NRG2_MOUSE
|
||||||
| NC score | 0.054737 (rank : 80) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P56974 | Gene names | Nrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Divergent of neuregulin 1) (DON-1)]. | |||||
|
NRG2_HUMAN
|
||||||
| NC score | 0.054325 (rank : 81) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14511 | Gene names | NRG2, NTAK | |||
|
Domain Architecture |

|
|||||
| Description | Pro-neuregulin-2, membrane-bound isoform precursor (Pro-NRG2) [Contains: Neuregulin-2 (NRG-2) (Neural- and thymus-derived activator for ERBB kinases) (NTAK) (Divergent of neuregulin-1) (DON-1)]. | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.054253 (rank : 82) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.054045 (rank : 83) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.053630 (rank : 84) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.052937 (rank : 85) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
MOX2R_MOUSE
|
||||||
| NC score | 0.052514 (rank : 86) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ES57 | Gene names | Cd200r1, Mox2r, Ox2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell surface glycoprotein OX2 receptor precursor (CD200 cell surface glycoprotein receptor). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.052271 (rank : 87) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.051893 (rank : 88) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
PUNC_HUMAN
|
||||||
| NC score | 0.051235 (rank : 89) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IVU1, O95215 | Gene names | PUNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.051132 (rank : 90) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.050474 (rank : 91) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
PUNC_MOUSE
|
||||||
| NC score | 0.050418 (rank : 92) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BQC3, O70246, Q792T2, Q9Z2S6 | Gene names | Punc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative neuronal cell adhesion molecule precursor. | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.040297 (rank : 93) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.039379 (rank : 94) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.038305 (rank : 95) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.037396 (rank : 96) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.033452 (rank : 97) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.032208 (rank : 98) | θ value | 0.47712 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.032208 (rank : 99) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.031055 (rank : 100) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
SO5A1_HUMAN
|
||||||
| NC score | 0.030971 (rank : 101) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H2Y9 | Gene names | SLCO5A1, SLC21A15 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 5A1 (Solute carrier family 21 member 15) (Organic anion transporter polypeptide- related protein 4) (OATP-RP4) (OATPRP4). | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.029309 (rank : 102) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CD244_MOUSE
|
||||||
| NC score | 0.028764 (rank : 103) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07763, O88654, Q9JIE0 | Gene names | Cd244, 2b4, Nmrk | |||
|
Domain Architecture |

|
|||||
| Description | Natural killer cell receptor 2B4 precursor (NKR2B4) (NK cell type I receptor protein 2B4) (CD244 antigen) (Non MHC restricted killing associated). | |||||
|
SO1B1_HUMAN
|
||||||
| NC score | 0.024382 (rank : 104) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6L6, Q9NQ37, Q9UBF3, Q9UH89 | Gene names | SLCO1B1, LST1, OATP2, OATPC, SLC21A6 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1B1 (Solute carrier family 21 member 6) (Sodium-independent organic anion- transporting polypeptide 2) (OATP 2) (Liver-specific organic anion transporter 1) (LST-1) (OATP-C). | |||||
|
SO1B3_HUMAN
|
||||||
| NC score | 0.022881 (rank : 105) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NPD5 | Gene names | SLCO1B3, LST2, SLC21A8 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1B3 (Solute carrier family 21 member 8) (Organic anion transporter 8) (Organic anion-transporting polypeptide 8) (OATP8) (Liver-specific organic anion transporter 2) (LST-2). | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.022094 (rank : 106) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
ATS20_MOUSE
|
||||||
| NC score | 0.014628 (rank : 107) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59511 | Gene names | Adamts20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||