Please be patient as the page loads
|
TINAG_HUMAN
|
||||||
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
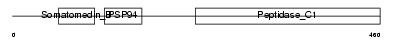
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TINAG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
TINAL_HUMAN
|
||||||
| θ value | 1.0968e-122 (rank : 2) | NC score | 0.973221 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 9.28496e-122 (rank : 3) | NC score | 0.971293 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
CATB_MOUSE
|
||||||
| θ value | 1.6247e-33 (rank : 4) | NC score | 0.878775 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10605, Q3UDW7 | Gene names | Ctsb | |||
|
Domain Architecture |
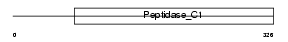
|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATB_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 5) | NC score | 0.875935 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATC_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 6) | NC score | 0.844610 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97821, O08853 | Gene names | Ctsc | |||
|
Domain Architecture |
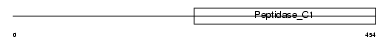
|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATC_HUMAN
|
||||||
| θ value | 4.14116e-29 (rank : 7) | NC score | 0.842168 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53634, Q8WYA7, Q8WYA8 | Gene names | CTSC, CPPI | |||
|
Domain Architecture |
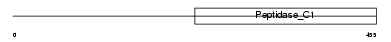
|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATL2_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 8) | NC score | 0.736073 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60911, O60233, Q5T1U0 | Gene names | CTSL2, CATL2, CTSU, CTSV | |||
|
Domain Architecture |
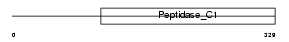
|
|||||
| Description | Cathepsin L2 precursor (EC 3.4.22.43) (Cathepsin V) (Cathepsin U). | |||||
|
CATH_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 9) | NC score | 0.774236 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49935 | Gene names | Ctsh | |||
|
Domain Architecture |
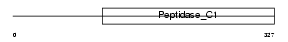
|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATZ_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 10) | NC score | 0.792868 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBR2, O75331, Q9UQV5, Q9UQV6 | Gene names | CTSZ | |||
|
Domain Architecture |
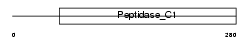
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P). | |||||
|
CATK_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 11) | NC score | 0.737689 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43235 | Gene names | CTSK, CTSO, CTSO2 | |||
|
Domain Architecture |
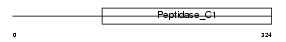
|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38) (Cathepsin O) (Cathepsin X) (Cathepsin O2). | |||||
|
CATK_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 12) | NC score | 0.742070 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55097, O88718 | Gene names | Ctsk | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38). | |||||
|
CATH_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 13) | NC score | 0.768306 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09668, Q9BUM7 | Gene names | CTSH | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATZ_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 14) | NC score | 0.788863 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CATS_HUMAN
|
||||||
| θ value | 1.29331e-14 (rank : 15) | NC score | 0.749996 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25774, Q5T5I0, Q9BUG3 | Gene names | CTSS | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATL_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 16) | NC score | 0.728169 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06797, Q91UZ0 | Gene names | Ctsl | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) (p39 cysteine proteinase) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATS_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 17) | NC score | 0.742731 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70370, O54973 | Gene names | Ctss, Cats | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATL_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 18) | NC score | 0.729423 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07711, Q96QJ0 | Gene names | CTSL | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATJ_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 19) | NC score | 0.733328 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R014, Q564D9, Q91XK6, Q9WV51 | Gene names | Ctsj, Ctsp | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin J precursor (EC 3.4.22.-) (Cathepsin P) (Cathepsin L-related protein) (Catlrp-p). | |||||
|
CATR_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 20) | NC score | 0.728183 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIA9 | Gene names | Ctsr, Catr | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin R precursor (EC 3.4.22.-). | |||||
|
CATO_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 21) | NC score | 0.739146 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43234 | Gene names | CTSO, CTSO1 | |||
|
Domain Architecture |
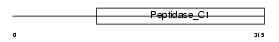
|
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATM_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 22) | NC score | 0.706065 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL96, Q91Z75, Q91ZF3, Q9CQB9 | Gene names | Ctsm, Catm | |||
|
Domain Architecture |
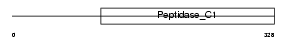
|
|||||
| Description | Cathepsin M precursor (EC 3.4.22.-). | |||||
|
CATW_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 23) | NC score | 0.714725 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56202, Q86VT4 | Gene names | CTSW | |||
|
Domain Architecture |
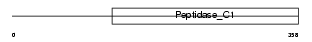
|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATW_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 24) | NC score | 0.720919 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P56203 | Gene names | Ctsw | |||
|
Domain Architecture |
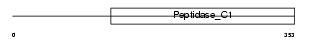
|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATO_MOUSE
|
||||||
| θ value | 2.13673e-09 (rank : 25) | NC score | 0.730641 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BM88, Q3TM69, Q80V35, Q8BKX0 | Gene names | Ctso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATF_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 26) | NC score | 0.686294 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBX1, O95240, Q9NSU4, Q9UKQ5 | Gene names | CTSF | |||
|
Domain Architecture |
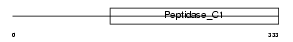
|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41) (CATSF). | |||||
|
VWF_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 27) | NC score | 0.152034 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 28) | NC score | 0.149922 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CATF_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 29) | NC score | 0.683766 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R013, Q9WUT4 | Gene names | Ctsf | |||
|
Domain Architecture |
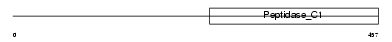
|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 30) | NC score | 0.106195 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 31) | NC score | 0.118883 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 32) | NC score | 0.136786 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 33) | NC score | 0.094474 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 34) | NC score | 0.087795 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.086801 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.071069 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
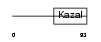
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.049948 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.070387 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
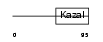
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
BMPER_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.119727 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.083087 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
FA10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.011274 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |
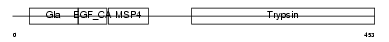
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.038449 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.080230 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.034774 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.034457 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
WISP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.037351 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |
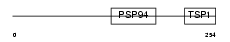
|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.104057 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.028694 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
JAG2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.026497 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.022217 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.031999 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.042381 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GALM_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.025779 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96C23, Q8NIA2 | Gene names | GALM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldose 1-epimerase (EC 5.1.3.3) (Galactose mutarotase) (BLOCK25 protein). | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.042515 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.069237 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TSP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.020224 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
VLDLR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.030037 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.031503 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
WISP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.032812 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
FA10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.008146 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |
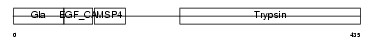
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.032980 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.025400 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
RECK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.046136 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.069616 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
CTL2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.381962 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12399 | Gene names | Ctla2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTLA-2-alpha protein precursor. | |||||
|
TECTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.072885 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.073256 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TSP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.241145 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13438 | Gene names | Tpbpa, Tpbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophoblast-specific protein alpha precursor. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.070824 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
TINAL_HUMAN
|
||||||
| NC score | 0.973221 (rank : 2) | θ value | 1.0968e-122 (rank : 2) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.971293 (rank : 3) | θ value | 9.28496e-122 (rank : 3) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
CATB_MOUSE
|
||||||
| NC score | 0.878775 (rank : 4) | θ value | 1.6247e-33 (rank : 4) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10605, Q3UDW7 | Gene names | Ctsb | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATB_HUMAN
|
||||||
| NC score | 0.875935 (rank : 5) | θ value | 3.06405e-32 (rank : 5) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATC_MOUSE
|
||||||
| NC score | 0.844610 (rank : 6) | θ value | 3.74554e-30 (rank : 6) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97821, O08853 | Gene names | Ctsc | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATC_HUMAN
|
||||||
| NC score | 0.842168 (rank : 7) | θ value | 4.14116e-29 (rank : 7) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53634, Q8WYA7, Q8WYA8 | Gene names | CTSC, CPPI | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATZ_HUMAN
|
||||||
| NC score | 0.792868 (rank : 8) | θ value | 1.38178e-16 (rank : 10) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBR2, O75331, Q9UQV5, Q9UQV6 | Gene names | CTSZ | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P). | |||||
|
CATZ_MOUSE
|
||||||
| NC score | 0.788863 (rank : 9) | θ value | 5.8054e-15 (rank : 14) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CATH_MOUSE
|
||||||
| NC score | 0.774236 (rank : 10) | θ value | 3.63628e-17 (rank : 9) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49935 | Gene names | Ctsh | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATH_HUMAN
|
||||||
| NC score | 0.768306 (rank : 11) | θ value | 2.60593e-15 (rank : 13) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09668, Q9BUM7 | Gene names | CTSH | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATS_HUMAN
|
||||||
| NC score | 0.749996 (rank : 12) | θ value | 1.29331e-14 (rank : 15) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25774, Q5T5I0, Q9BUG3 | Gene names | CTSS | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATS_MOUSE
|
||||||
| NC score | 0.742731 (rank : 13) | θ value | 8.38298e-14 (rank : 17) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70370, O54973 | Gene names | Ctss, Cats | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATK_MOUSE
|
||||||
| NC score | 0.742070 (rank : 14) | θ value | 1.99529e-15 (rank : 12) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55097, O88718 | Gene names | Ctsk | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38). | |||||
|
CATO_HUMAN
|
||||||
| NC score | 0.739146 (rank : 15) | θ value | 3.52202e-12 (rank : 21) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43234 | Gene names | CTSO, CTSO1 | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATK_HUMAN
|
||||||
| NC score | 0.737689 (rank : 16) | θ value | 8.95645e-16 (rank : 11) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43235 | Gene names | CTSK, CTSO, CTSO2 | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38) (Cathepsin O) (Cathepsin X) (Cathepsin O2). | |||||
|
CATL2_HUMAN
|
||||||
| NC score | 0.736073 (rank : 17) | θ value | 2.7842e-17 (rank : 8) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60911, O60233, Q5T1U0 | Gene names | CTSL2, CATL2, CTSU, CTSV | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L2 precursor (EC 3.4.22.43) (Cathepsin V) (Cathepsin U). | |||||
|
CATJ_MOUSE
|
||||||
| NC score | 0.733328 (rank : 18) | θ value | 2.43908e-13 (rank : 19) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R014, Q564D9, Q91XK6, Q9WV51 | Gene names | Ctsj, Ctsp | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin J precursor (EC 3.4.22.-) (Cathepsin P) (Cathepsin L-related protein) (Catlrp-p). | |||||
|
CATO_MOUSE
|
||||||
| NC score | 0.730641 (rank : 19) | θ value | 2.13673e-09 (rank : 25) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BM88, Q3TM69, Q80V35, Q8BKX0 | Gene names | Ctso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATL_HUMAN
|
||||||
| NC score | 0.729423 (rank : 20) | θ value | 1.09485e-13 (rank : 18) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07711, Q96QJ0 | Gene names | CTSL | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATR_MOUSE
|
||||||
| NC score | 0.728183 (rank : 21) | θ value | 2.43908e-13 (rank : 20) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIA9 | Gene names | Ctsr, Catr | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin R precursor (EC 3.4.22.-). | |||||
|
CATL_MOUSE
|
||||||
| NC score | 0.728169 (rank : 22) | θ value | 2.20605e-14 (rank : 16) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06797, Q91UZ0 | Gene names | Ctsl | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) (p39 cysteine proteinase) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATW_MOUSE
|
||||||
| NC score | 0.720919 (rank : 23) | θ value | 7.34386e-10 (rank : 24) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P56203 | Gene names | Ctsw | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATW_HUMAN
|
||||||
| NC score | 0.714725 (rank : 24) | θ value | 1.9326e-10 (rank : 23) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56202, Q86VT4 | Gene names | CTSW | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATM_MOUSE
|
||||||
| NC score | 0.706065 (rank : 25) | θ value | 4.59992e-12 (rank : 22) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL96, Q91Z75, Q91ZF3, Q9CQB9 | Gene names | Ctsm, Catm | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin M precursor (EC 3.4.22.-). | |||||
|
CATF_HUMAN
|
||||||
| NC score | 0.686294 (rank : 26) | θ value | 4.92598e-06 (rank : 26) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBX1, O95240, Q9NSU4, Q9UKQ5 | Gene names | CTSF | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41) (CATSF). | |||||
|
CATF_MOUSE
|
||||||
| NC score | 0.683766 (rank : 27) | θ value | 0.000121331 (rank : 29) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R013, Q9WUT4 | Gene names | Ctsf | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41). | |||||
|
CTL2A_MOUSE
|
||||||
| NC score | 0.381962 (rank : 28) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12399 | Gene names | Ctla2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTLA-2-alpha protein precursor. | |||||
|
TSP_MOUSE
|
||||||
| NC score | 0.241145 (rank : 29) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13438 | Gene names | Tpbpa, Tpbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophoblast-specific protein alpha precursor. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.152034 (rank : 30) | θ value | 2.44474e-05 (rank : 27) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.149922 (rank : 31) | θ value | 5.44631e-05 (rank : 28) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.136786 (rank : 32) | θ value | 0.000602161 (rank : 32) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.119727 (rank : 33) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.118883 (rank : 34) | θ value | 0.000602161 (rank : 31) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.106195 (rank : 35) | θ value | 0.000461057 (rank : 30) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.104057 (rank : 36) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.094474 (rank : 37) | θ value | 0.00134147 (rank : 33) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.087795 (rank : 38) | θ value | 0.0330416 (rank : 34) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.086801 (rank : 39) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.083087 (rank : 40) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.080230 (rank : 41) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.073256 (rank : 42) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.072885 (rank : 43) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.071069 (rank : 44) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.070824 (rank : 45) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.070387 (rank : 46) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.069616 (rank : 47) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.069237 (rank : 48) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.049948 (rank : 49) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.046136 (rank : 50) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.042515 (rank : 51) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.042381 (rank : 52) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.038449 (rank : 53) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
WISP1_HUMAN
|
||||||
| NC score | 0.037351 (rank : 54) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.034774 (rank : 55) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.034457 (rank : 56) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.032980 (rank : 57) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
WISP1_MOUSE
|
||||||
| NC score | 0.032812 (rank : 58) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.031999 (rank : 59) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.031503 (rank : 60) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
VLDLR_MOUSE
|
||||||
| NC score | 0.030037 (rank : 61) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98156, Q64022 | Gene names | Vldlr | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.028694 (rank : 62) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.026497 (rank : 63) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
GALM_HUMAN
|
||||||
| NC score | 0.025779 (rank : 64) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96C23, Q8NIA2 | Gene names | GALM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Aldose 1-epimerase (EC 5.1.3.3) (Galactose mutarotase) (BLOCK25 protein). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.025400 (rank : 65) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.022217 (rank : 66) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
TSP2_HUMAN
|
||||||
| NC score | 0.020224 (rank : 67) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
FA10_MOUSE
|
||||||
| NC score | 0.011274 (rank : 68) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA10_HUMAN
|
||||||
| NC score | 0.008146 (rank : 69) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 63 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||