Please be patient as the page loads
|
TINAL_HUMAN
|
||||||
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TINAL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998088 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
TINAG_HUMAN
|
||||||
| θ value | 1.0968e-122 (rank : 3) | NC score | 0.973221 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
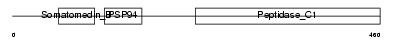
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
CATB_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 4) | NC score | 0.887960 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATB_MOUSE
|
||||||
| θ value | 1.6774e-38 (rank : 5) | NC score | 0.890654 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10605, Q3UDW7 | Gene names | Ctsb | |||
|
Domain Architecture |
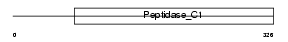
|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATC_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 6) | NC score | 0.845016 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97821, O08853 | Gene names | Ctsc | |||
|
Domain Architecture |
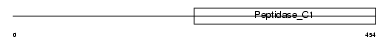
|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATC_HUMAN
|
||||||
| θ value | 9.2256e-29 (rank : 7) | NC score | 0.843078 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53634, Q8WYA7, Q8WYA8 | Gene names | CTSC, CPPI | |||
|
Domain Architecture |
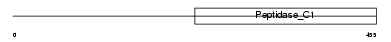
|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATH_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 8) | NC score | 0.780065 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49935 | Gene names | Ctsh | |||
|
Domain Architecture |
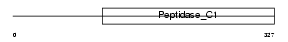
|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATH_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 9) | NC score | 0.774528 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09668, Q9BUM7 | Gene names | CTSH | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATS_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 10) | NC score | 0.754159 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25774, Q5T5I0, Q9BUG3 | Gene names | CTSS | |||
|
Domain Architecture |
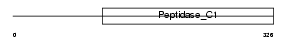
|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATJ_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 11) | NC score | 0.737760 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R014, Q564D9, Q91XK6, Q9WV51 | Gene names | Ctsj, Ctsp | |||
|
Domain Architecture |
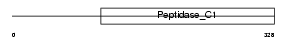
|
|||||
| Description | Cathepsin J precursor (EC 3.4.22.-) (Cathepsin P) (Cathepsin L-related protein) (Catlrp-p). | |||||
|
CATR_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 12) | NC score | 0.732434 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIA9 | Gene names | Ctsr, Catr | |||
|
Domain Architecture |
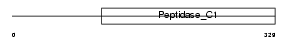
|
|||||
| Description | Cathepsin R precursor (EC 3.4.22.-). | |||||
|
CATS_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 13) | NC score | 0.746449 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70370, O54973 | Gene names | Ctss, Cats | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATL2_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 14) | NC score | 0.736758 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60911, O60233, Q5T1U0 | Gene names | CTSL2, CATL2, CTSU, CTSV | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L2 precursor (EC 3.4.22.43) (Cathepsin V) (Cathepsin U). | |||||
|
CATK_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 15) | NC score | 0.743276 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55097, O88718 | Gene names | Ctsk | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38). | |||||
|
CATL_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 16) | NC score | 0.732600 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07711, Q96QJ0 | Gene names | CTSL | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATZ_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 17) | NC score | 0.787088 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBR2, O75331, Q9UQV5, Q9UQV6 | Gene names | CTSZ | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P). | |||||
|
CATL_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 18) | NC score | 0.731049 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06797, Q91UZ0 | Gene names | Ctsl | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) (p39 cysteine proteinase) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATZ_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 19) | NC score | 0.784426 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CATW_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 20) | NC score | 0.732532 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56203 | Gene names | Ctsw | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATK_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 21) | NC score | 0.736459 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43235 | Gene names | CTSK, CTSO, CTSO2 | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38) (Cathepsin O) (Cathepsin X) (Cathepsin O2). | |||||
|
CATW_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 22) | NC score | 0.724492 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56202, Q86VT4 | Gene names | CTSW | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATM_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 23) | NC score | 0.707375 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL96, Q91Z75, Q91ZF3, Q9CQB9 | Gene names | Ctsm, Catm | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin M precursor (EC 3.4.22.-). | |||||
|
CATO_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 24) | NC score | 0.738413 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43234 | Gene names | CTSO, CTSO1 | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATO_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 25) | NC score | 0.732051 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BM88, Q3TM69, Q80V35, Q8BKX0 | Gene names | Ctso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATF_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 26) | NC score | 0.700414 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBX1, O95240, Q9NSU4, Q9UKQ5 | Gene names | CTSF | |||
|
Domain Architecture |
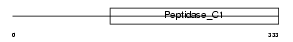
|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41) (CATSF). | |||||
|
CATF_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 27) | NC score | 0.698426 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R013, Q9WUT4 | Gene names | Ctsf | |||
|
Domain Architecture |
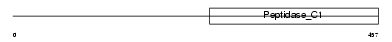
|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41). | |||||
|
VWF_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 28) | NC score | 0.119999 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 29) | NC score | 0.121630 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 30) | NC score | 0.076621 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.102689 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
BMPER_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.102161 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
NRDC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 33) | NC score | 0.055768 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.063673 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.030133 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.058469 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
NELL2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.024293 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NRDC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.043365 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
BMPER_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.083497 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.084140 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.018981 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.051008 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.013612 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.014338 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.018422 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.013371 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.013945 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
TECTA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.056667 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.056506 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
DLL4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.014343 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
CTL2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.379267 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12399 | Gene names | Ctla2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTLA-2-alpha protein precursor. | |||||
|
TSP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.237916 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13438 | Gene names | Tpbpa, Tpbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophoblast-specific protein alpha precursor. | |||||
|
ZAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.051958 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TINAL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9GZM7, Q8TEJ9, Q8WZ23, Q96GZ4, Q96JW3 | Gene names | TINAGL1, GIS5, LCN7, OLRG2, TINAGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Tubulointerstitial nephritis antigen-related protein) (TIN Ag-related protein) (TIN-Ag-RP) (Glucocorticoid-inducible protein 5) (Oxidized LDL-responsive gene 2 protein) (OLRG-2). | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.998088 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.973221 (rank : 3) | θ value | 1.0968e-122 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
CATB_MOUSE
|
||||||
| NC score | 0.890654 (rank : 4) | θ value | 1.6774e-38 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10605, Q3UDW7 | Gene names | Ctsb | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATB_HUMAN
|
||||||
| NC score | 0.887960 (rank : 5) | θ value | 9.83387e-39 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07858, Q96D87 | Gene names | CTSB, CPSB | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin B precursor (EC 3.4.22.1) (Cathepsin B1) (APP secretase) (APPS) [Contains: Cathepsin B light chain; Cathepsin B heavy chain]. | |||||
|
CATC_MOUSE
|
||||||
| NC score | 0.845016 (rank : 6) | θ value | 5.40856e-29 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97821, O08853 | Gene names | Ctsc | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATC_HUMAN
|
||||||
| NC score | 0.843078 (rank : 7) | θ value | 9.2256e-29 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P53634, Q8WYA7, Q8WYA8 | Gene names | CTSC, CPPI | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl-peptidase 1 precursor (EC 3.4.14.1) (Dipeptidyl-peptidase I) (DPP-I) (DPPI) (Cathepsin C) (Cathepsin J) (Dipeptidyl transferase) [Contains: Dipeptidyl-peptidase 1 exclusion domain chain (Dipeptidyl- peptidase I exclusion domain chain); Dipeptidyl-peptidase 1 heavy chain (Dipeptidyl-peptidase I heavy chain); Dipeptidyl-peptidase 1 light chain (Dipeptidyl-peptidase I light chain)]. | |||||
|
CATZ_HUMAN
|
||||||
| NC score | 0.787088 (rank : 8) | θ value | 9.90251e-15 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBR2, O75331, Q9UQV5, Q9UQV6 | Gene names | CTSZ | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-) (Cathepsin X) (Cathepsin P). | |||||
|
CATZ_MOUSE
|
||||||
| NC score | 0.784426 (rank : 9) | θ value | 2.20605e-14 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WUU7 | Gene names | Ctsz | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin Z precursor (EC 3.4.22.-). | |||||
|
CATH_MOUSE
|
||||||
| NC score | 0.780065 (rank : 10) | θ value | 3.88503e-19 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49935 | Gene names | Ctsh | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) (Cathepsin B3) (Cathepsin BA) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATH_HUMAN
|
||||||
| NC score | 0.774528 (rank : 11) | θ value | 1.63225e-17 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09668, Q9BUM7 | Gene names | CTSH | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin H precursor (EC 3.4.22.16) [Contains: Cathepsin H mini chain; Cathepsin H heavy chain; Cathepsin H light chain]. | |||||
|
CATS_HUMAN
|
||||||
| NC score | 0.754159 (rank : 12) | θ value | 2.13179e-17 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25774, Q5T5I0, Q9BUG3 | Gene names | CTSS | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATS_MOUSE
|
||||||
| NC score | 0.746449 (rank : 13) | θ value | 4.02038e-16 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70370, O54973 | Gene names | Ctss, Cats | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin S precursor (EC 3.4.22.27). | |||||
|
CATK_MOUSE
|
||||||
| NC score | 0.743276 (rank : 14) | θ value | 2.60593e-15 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55097, O88718 | Gene names | Ctsk | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38). | |||||
|
CATO_HUMAN
|
||||||
| NC score | 0.738413 (rank : 15) | θ value | 5.62301e-10 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43234 | Gene names | CTSO, CTSO1 | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATJ_MOUSE
|
||||||
| NC score | 0.737760 (rank : 16) | θ value | 1.058e-16 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R014, Q564D9, Q91XK6, Q9WV51 | Gene names | Ctsj, Ctsp | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin J precursor (EC 3.4.22.-) (Cathepsin P) (Cathepsin L-related protein) (Catlrp-p). | |||||
|
CATL2_HUMAN
|
||||||
| NC score | 0.736758 (rank : 17) | θ value | 1.16975e-15 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60911, O60233, Q5T1U0 | Gene names | CTSL2, CATL2, CTSU, CTSV | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L2 precursor (EC 3.4.22.43) (Cathepsin V) (Cathepsin U). | |||||
|
CATK_HUMAN
|
||||||
| NC score | 0.736459 (rank : 18) | θ value | 7.84624e-12 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P43235 | Gene names | CTSK, CTSO, CTSO2 | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin K precursor (EC 3.4.22.38) (Cathepsin O) (Cathepsin X) (Cathepsin O2). | |||||
|
CATL_HUMAN
|
||||||
| NC score | 0.732600 (rank : 19) | θ value | 2.60593e-15 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07711, Q96QJ0 | Gene names | CTSL | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATW_MOUSE
|
||||||
| NC score | 0.732532 (rank : 20) | θ value | 1.2105e-12 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56203 | Gene names | Ctsw | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATR_MOUSE
|
||||||
| NC score | 0.732434 (rank : 21) | θ value | 1.80466e-16 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIA9 | Gene names | Ctsr, Catr | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin R precursor (EC 3.4.22.-). | |||||
|
CATO_MOUSE
|
||||||
| NC score | 0.732051 (rank : 22) | θ value | 5.62301e-10 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BM88, Q3TM69, Q80V35, Q8BKX0 | Gene names | Ctso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cathepsin O precursor (EC 3.4.22.42). | |||||
|
CATL_MOUSE
|
||||||
| NC score | 0.731049 (rank : 23) | θ value | 1.68911e-14 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P06797, Q91UZ0 | Gene names | Ctsl | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin L precursor (EC 3.4.22.15) (Major excreted protein) (MEP) (p39 cysteine proteinase) [Contains: Cathepsin L heavy chain; Cathepsin L light chain]. | |||||
|
CATW_HUMAN
|
||||||
| NC score | 0.724492 (rank : 24) | θ value | 3.89403e-11 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P56202, Q86VT4 | Gene names | CTSW | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin W precursor (EC 3.4.22.-) (Lymphopain). | |||||
|
CATM_MOUSE
|
||||||
| NC score | 0.707375 (rank : 25) | θ value | 5.08577e-11 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JL96, Q91Z75, Q91ZF3, Q9CQB9 | Gene names | Ctsm, Catm | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin M precursor (EC 3.4.22.-). | |||||
|
CATF_HUMAN
|
||||||
| NC score | 0.700414 (rank : 26) | θ value | 1.38499e-08 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UBX1, O95240, Q9NSU4, Q9UKQ5 | Gene names | CTSF | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41) (CATSF). | |||||
|
CATF_MOUSE
|
||||||
| NC score | 0.698426 (rank : 27) | θ value | 2.36244e-08 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R013, Q9WUT4 | Gene names | Ctsf | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin F precursor (EC 3.4.22.41). | |||||
|
CTL2A_MOUSE
|
||||||
| NC score | 0.379267 (rank : 28) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12399 | Gene names | Ctla2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTLA-2-alpha protein precursor. | |||||
|
TSP_MOUSE
|
||||||
| NC score | 0.237916 (rank : 29) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P13438 | Gene names | Tpbpa, Tpbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trophoblast-specific protein alpha precursor. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.121630 (rank : 30) | θ value | 0.000786445 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.119999 (rank : 31) | θ value | 0.00035302 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.102689 (rank : 32) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
BMPER_HUMAN
|
||||||
| NC score | 0.102161 (rank : 33) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8N8U9 | Gene names | BMPER | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (hCV2). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.084140 (rank : 34) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
BMPER_MOUSE
|
||||||
| NC score | 0.083497 (rank : 35) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CJ69, Q7TN57, Q80UZ1, Q9CXM8 | Gene names | Bmper, Cv2 | |||
|
Domain Architecture |

|
|||||
| Description | BMP-binding endothelial regulator protein precursor (Bone morphogenetic protein-binding endothelial cell precursor-derived regulator) (Protein crossveinless-2) (mCV2). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.076621 (rank : 36) | θ value | 0.00298849 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.063673 (rank : 37) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.058469 (rank : 38) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.056667 (rank : 39) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.056506 (rank : 40) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
NRDC_HUMAN
|
||||||
| NC score | 0.055768 (rank : 41) | θ value | 0.163984 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43847, O15241, O15242, Q5VUL0, Q96HB2, Q9NU57 | Gene names | NRD1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.051958 (rank : 42) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.051008 (rank : 43) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
NRDC_MOUSE
|
||||||
| NC score | 0.043365 (rank : 44) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHG1 | Gene names | Nrd1 | |||
|
Domain Architecture |

|
|||||
| Description | Nardilysin precursor (EC 3.4.24.61) (N-arginine dibasic convertase) (NRD convertase) (NRD-C). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.030133 (rank : 45) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.024293 (rank : 46) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.018981 (rank : 47) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.018422 (rank : 48) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.014343 (rank : 49) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.014338 (rank : 50) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.013945 (rank : 51) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.013612 (rank : 52) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.013371 (rank : 53) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||