Please be patient as the page loads
|
SMOC2_MOUSE
|
||||||
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
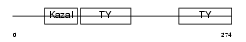
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMOC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996339 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
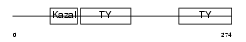
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | 2.97994e-128 (rank : 3) | NC score | 0.974760 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
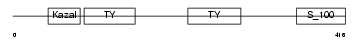
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 1.80787e-125 (rank : 4) | NC score | 0.972557 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
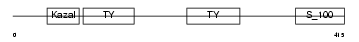
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 5) | NC score | 0.278386 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID2_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 6) | NC score | 0.278918 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
THYG_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 7) | NC score | 0.369060 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 8) | NC score | 0.352299 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 9) | NC score | 0.527447 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
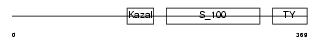
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
NID1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 10) | NC score | 0.247822 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 11) | NC score | 0.524830 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
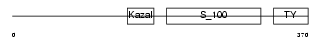
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
NID1_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 12) | NC score | 0.243673 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
HG2A_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 13) | NC score | 0.537840 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
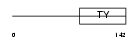
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 14) | NC score | 0.491329 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
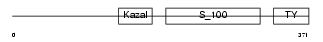
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 15) | NC score | 0.489315 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 16) | NC score | 0.379833 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
HG2A_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 17) | NC score | 0.492474 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
FST_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 18) | NC score | 0.355787 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.348472 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 20) | NC score | 0.403707 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
TICN3_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 21) | NC score | 0.478900 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
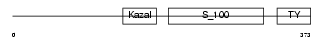
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 22) | NC score | 0.297494 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 23) | NC score | 0.297292 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 24) | NC score | 0.476554 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
SPRC_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 25) | NC score | 0.359444 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
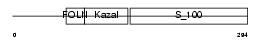
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 26) | NC score | 0.399906 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
SPRC_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 27) | NC score | 0.357769 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
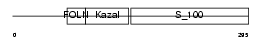
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 28) | NC score | 0.164703 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 29) | NC score | 0.376611 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 30) | NC score | 0.282080 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 31) | NC score | 0.337345 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 32) | NC score | 0.384020 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
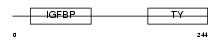
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 33) | NC score | 0.386066 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
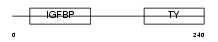
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 34) | NC score | 0.305558 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 35) | NC score | 0.366622 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 36) | NC score | 0.350997 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
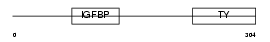
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 37) | NC score | 0.385022 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 38) | NC score | 0.368461 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
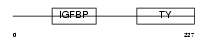
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 39) | NC score | 0.310884 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 40) | NC score | 0.162070 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.270459 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
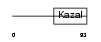
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.270033 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 43) | NC score | 0.165535 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 44) | NC score | 0.337402 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.108316 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.071115 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.334217 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
TACD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.289444 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.030604 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
FSTL5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.108768 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
ISK6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.202337 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.034354 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.058198 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.014385 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.057089 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
SO1A2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.020374 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46721, Q9UGP7, Q9UL38 | Gene names | SLCO1A2, OATP, SLC21A3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1A2 (Solute carrier family 21 member 3) (Sodium-independent organic anion transporter) (Organic anion-transporting polypeptide 1) (OATP1) (OATP- A). | |||||
|
TACD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.240177 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
KCIP4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.055928 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.055914 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
CSEN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.053442 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.026905 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
TULP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.023394 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.121098 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
S10A9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.032737 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06702, Q9NYM0, Q9UCJ1 | Gene names | S100A9, CAGB, MRP14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain) (Calprotectin L1H subunit). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.101504 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.129941 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.023160 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
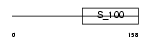
GUC1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.050982 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.046018 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.046018 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.101106 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
KCIP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.051351 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.051275 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.101293 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.047948 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
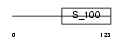
HPCL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.043228 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.043535 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
PASK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | -0.002349 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||
|
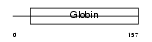
CYGB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.014546 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CX80 | Gene names | Cygb | |||
|
Domain Architecture |
|
|||||
| Description | Cytoglobin (Histoglobin) (HGb). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.016819 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PKP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.010955 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
EGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055144 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
EGF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.055070 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.085455 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
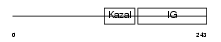
IBP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.085878 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.079918 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
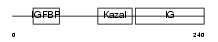
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
IPK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.082437 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
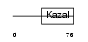
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
IPK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.061284 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |
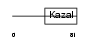
|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.059559 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
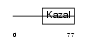
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
ISK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.095810 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |
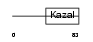
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
ISK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.100355 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |
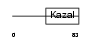
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
ISK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.072625 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |
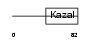
|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
ISK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.073569 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
LRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.059216 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.058955 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
LRP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.057309 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.057247 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
RECK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051794 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
TECTA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.053558 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
TECTA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.051438 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.996339 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.974760 (rank : 3) | θ value | 2.97994e-128 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.972557 (rank : 4) | θ value | 1.80787e-125 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.537840 (rank : 5) | θ value | 7.59969e-07 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.527447 (rank : 6) | θ value | 3.64472e-09 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.524830 (rank : 7) | θ value | 5.26297e-08 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
HG2A_HUMAN
|
||||||
| NC score | 0.492474 (rank : 8) | θ value | 1.09739e-05 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.491329 (rank : 9) | θ value | 2.21117e-06 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.489315 (rank : 10) | θ value | 2.21117e-06 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.478900 (rank : 11) | θ value | 4.1701e-05 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.476554 (rank : 12) | θ value | 5.44631e-05 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.403707 (rank : 13) | θ value | 4.1701e-05 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.399906 (rank : 14) | θ value | 0.000121331 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.386066 (rank : 15) | θ value | 0.000461057 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.385022 (rank : 16) | θ value | 0.00509761 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.384020 (rank : 17) | θ value | 0.000461057 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.379833 (rank : 18) | θ value | 8.40245e-06 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.376611 (rank : 19) | θ value | 0.00035302 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.369060 (rank : 20) | θ value | 2.60593e-15 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.368461 (rank : 21) | θ value | 0.00665767 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.366622 (rank : 22) | θ value | 0.00134147 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.359444 (rank : 23) | θ value | 9.29e-05 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.357769 (rank : 24) | θ value | 0.000158464 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.355787 (rank : 25) | θ value | 1.43324e-05 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.352299 (rank : 26) | θ value | 1.09485e-13 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.350997 (rank : 27) | θ value | 0.00390308 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.348472 (rank : 28) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.337402 (rank : 29) | θ value | 0.0563607 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.337345 (rank : 30) | θ value | 0.000461057 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.334217 (rank : 31) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.310884 (rank : 32) | θ value | 0.0148317 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.305558 (rank : 33) | θ value | 0.000786445 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.297494 (rank : 34) | θ value | 5.44631e-05 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.297292 (rank : 35) | θ value | 5.44631e-05 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.289444 (rank : 36) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.282080 (rank : 37) | θ value | 0.00035302 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.278918 (rank : 38) | θ value | 1.99529e-15 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.278386 (rank : 39) | θ value | 1.99529e-15 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.270459 (rank : 40) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.270033 (rank : 41) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.247822 (rank : 42) | θ value | 1.80886e-08 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.243673 (rank : 43) | θ value | 6.87365e-08 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
TACD2_HUMAN
|
||||||
| NC score | 0.240177 (rank : 44) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
ISK6_MOUSE
|
||||||
| NC score | 0.202337 (rank : 45) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.165535 (rank : 46) | θ value | 0.0431538 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.164703 (rank : 47) | θ value | 0.00020696 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.162070 (rank : 48) | θ value | 0.0193708 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.129941 (rank : 49) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.121098 (rank : 50) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
FSTL5_HUMAN
|
||||||
| NC score | 0.108768 (rank : 51) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.108316 (rank : 52) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.101504 (rank : 53) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.101293 (rank : 54) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.101106 (rank : 55) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
ISK4_MOUSE
|
||||||
| NC score | 0.100355 (rank : 56) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
ISK4_HUMAN
|
||||||
| NC score | 0.095810 (rank : 57) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.085878 (rank : 58) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.085455 (rank : 59) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.082437 (rank : 60) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.079918 (rank : 61) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
ISK7_MOUSE
|
||||||
| NC score | 0.073569 (rank : 62) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
ISK7_HUMAN
|
||||||
| NC score | 0.072625 (rank : 63) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.071115 (rank : 64) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
IPK2_HUMAN
|
||||||
| NC score | 0.061284 (rank : 65) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.059559 (rank : 66) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
LRP5_HUMAN
|
||||||
| NC score | 0.059216 (rank : 67) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75197, Q96TD6, Q9UP66 | Gene names | LRP5, LRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor. | |||||
|
LRP5_MOUSE
|
||||||
| NC score | 0.058955 (rank : 68) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.058198 (rank : 69) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
LRP6_HUMAN
|
||||||
| NC score | 0.057309 (rank : 70) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75581 | Gene names | LRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
LRP6_MOUSE
|
||||||
| NC score | 0.057247 (rank : 71) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88572 | Gene names | Lrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 6 precursor. | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.057089 (rank : 72) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP4_HUMAN
|
||||||
| NC score | 0.055928 (rank : 73) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| NC score | 0.055914 (rank : 74) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
EGF_HUMAN
|
||||||
| NC score | 0.055144 (rank : 75) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
EGF_MOUSE
|
||||||
| NC score | 0.055070 (rank : 76) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
TECTA_HUMAN
|
||||||
| NC score | 0.053558 (rank : 77) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75443 | Gene names | TECTA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.053442 (rank : 78) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.051794 (rank : 79) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.051438 (rank : 80) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
KCIP1_HUMAN
|
||||||
| NC score | 0.051351 (rank : 81) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| NC score | 0.051275 (rank : 82) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
GUC1A_MOUSE
|
||||||
| NC score | 0.050982 (rank : 83) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.047948 (rank : 84) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.046018 (rank : 85) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.046018 (rank : 86) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.043535 (rank : 87) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.043228 (rank : 88) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.034354 (rank : 89) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
S10A9_HUMAN
|
||||||
| NC score | 0.032737 (rank : 90) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06702, Q9NYM0, Q9UCJ1 | Gene names | S100A9, CAGB, MRP14 | |||
|
Domain Architecture |

|
|||||
| Description | Protein S100-A9 (S100 calcium-binding protein A9) (Calgranulin-B) (Migration inhibitory factor-related protein 14) (MRP-14) (P14) (Leukocyte L1 complex heavy chain) (Calprotectin L1H subunit). | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.030604 (rank : 91) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.026905 (rank : 92) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
TULP1_MOUSE
|
||||||
| NC score | 0.023394 (rank : 93) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z273 | Gene names | Tulp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.023160 (rank : 94) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
SO1A2_HUMAN
|
||||||
| NC score | 0.020374 (rank : 95) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46721, Q9UGP7, Q9UL38 | Gene names | SLCO1A2, OATP, SLC21A3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 1A2 (Solute carrier family 21 member 3) (Sodium-independent organic anion transporter) (Organic anion-transporting polypeptide 1) (OATP1) (OATP- A). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.016819 (rank : 96) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CYGB_MOUSE
|
||||||
| NC score | 0.014546 (rank : 97) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CX80 | Gene names | Cygb | |||
|
Domain Architecture |

|
|||||
| Description | Cytoglobin (Histoglobin) (HGb). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.014385 (rank : 98) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.010955 (rank : 99) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PASK_MOUSE
|
||||||
| NC score | -0.002349 (rank : 100) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CEE6, Q91YD8 | Gene names | Pask, Kiaa0135 | |||
|
Domain Architecture |

|
|||||
| Description | PAS domain-containing serine/threonine-protein kinase (EC 2.7.11.1) (PAS-kinase) (PASKIN). | |||||