Please be patient as the page loads
|
NUMB_HUMAN
|
||||||
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUMB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.983232 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 6.17048e-150 (rank : 3) | NC score | 0.850814 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 8.0589e-150 (rank : 4) | NC score | 0.937837 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
ARH_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 5) | NC score | 0.588098 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ARH_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 6) | NC score | 0.576376 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 7) | NC score | 0.390019 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 8) | NC score | 0.397747 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |
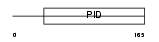
|
|||||
| Description | Disabled homolog 1. | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 9) | NC score | 0.157028 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
ANKS1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 10) | NC score | 0.165522 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 11) | NC score | 0.334251 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
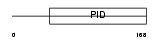
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 12) | NC score | 0.337659 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
CAPON_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 13) | NC score | 0.351611 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
CAPON_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 14) | NC score | 0.340742 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 15) | NC score | 0.213535 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.203664 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
SHC2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 17) | NC score | 0.110911 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMC3 | Gene names | Shc2, Sck, ShcB | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 2 (SH2 domain protein C2) (Src homology 2 domain-containing-transforming protein C2) (Protein Sck) (Protein Sli). | |||||
|
SHC2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.107710 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98077, O60230, Q9NPL5, Q9UCX4 | Gene names | SHC2, SCK, SHCB | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 2 (SH2 domain protein C2) (Src homology 2 domain-containing-transforming protein C2) (Protein Sck). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.017509 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.038505 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.122400 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.035188 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 23) | NC score | 0.038691 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
FA43A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.200841 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N2R8, Q8IXP4, Q8WZ07 | Gene names | FAM43A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
FA43A_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.200742 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUP8, Q8R231 | Gene names | Fam43a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
TM108_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.062510 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.056732 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.024513 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.056206 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.064535 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
HNF1A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.040799 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.033116 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SN1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.005383 (rank : 123) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57059, Q5R2V5, Q86YJ2 | Gene names | SNF1LK | |||
|
Domain Architecture |
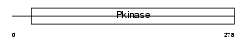
|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.045956 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.042296 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
APBB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.071348 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
FBX43_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.032452 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4G163 | Gene names | FBXO43, EMI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 43 (Endogenous meiotic inhibitor 2). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.060421 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.052453 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.037201 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.036569 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.056768 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CN032_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.072236 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.041291 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.047536 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.040992 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
B3A2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.012133 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04920, Q45EY5, Q969L3 | Gene names | SLC4A2, AE2, EPB3L1, HKB3, MPB3L | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 2 (Non-erythroid band 3-like protein) (AE2 anion exchanger) (Solute carrier family 4 member 2) (BND3L). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.050309 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.019844 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.011435 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.097242 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.042647 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.032783 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.047602 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.026092 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PER2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.054284 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.021271 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
APBB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.065290 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.055693 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.076326 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CT174_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.033956 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
DTX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.016849 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |
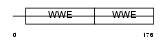
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.056324 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.020473 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.044443 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.028694 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.066998 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ADA29_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.014566 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.038732 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
HXD12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.006648 (rank : 122) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35452, Q9NS03 | Gene names | HOXD12, HOX4H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4H). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.009226 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.012850 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.014345 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.021794 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
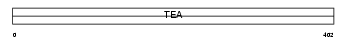
TEAD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.015289 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28347 | Gene names | TEAD1, TCF13, TEF1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcriptional enhancer factor TEF-1 (TEA domain family member 1) (TEAD-1) (Protein GT-IIC) (Transcription factor 13) (NTEF-1). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.040625 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
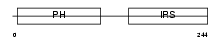
DOK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.018975 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
GSLG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.016667 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
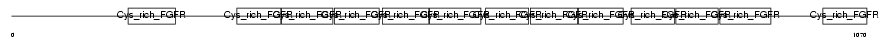
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.036783 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.011187 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.015642 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
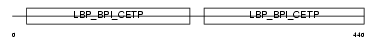
BPIL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.012956 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BU51, Q80ZU8 | Gene names | Bpil3 | |||
|
Domain Architecture |
|
|||||
| Description | Bactericidal/permeability-increasing protein-like 3 precursor. | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.033425 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.011349 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.007224 (rank : 121) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.055298 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRKN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.019981 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.026831 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
TLN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.007741 (rank : 120) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
WWP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.008409 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.033238 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.017783 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.050498 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.037140 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.009585 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
PXDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.010826 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.022350 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.037596 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TBCD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.027875 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60343, Q5W0B9 | Gene names | TBC1D4, KIAA0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
TEAD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.013017 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30051 | Gene names | Tead1, Tcf13, Tef-1, Tef1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcriptional enhancer factor TEF-1 (TEA domain family member 1) (TEAD-1) (Protein GT-IIC) (Transcription factor 13) (NTEF-1). | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.034974 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.023392 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.034950 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
GBX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.003320 (rank : 126) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14549 | Gene names | GBX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein GBX-1 (Gastrulation and brain-specific homeobox protein 1). | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.019500 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.004458 (rank : 125) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.013124 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.011525 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
PVRL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.004926 (rank : 124) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15223, O75465, Q9HBE6, Q9HBW2 | Gene names | PVRL1, HVEC, PRR1 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 1 precursor (Herpes virus entry mediator C) (HveC) (Nectin-1) (Herpesvirus Ig-like receptor) (HIgR) (CD111 antigen). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.009267 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
TBCD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.023155 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
TBCD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.028514 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.058004 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.075060 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
FA43B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.065991 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZT52 | Gene names | FAM43B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43B. | |||||
|
JIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.054294 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
JIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052371 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.058027 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.052779 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.056161 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.053253 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.060343 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.051346 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.050163 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.058907 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.080909 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.983232 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.937837 (rank : 3) | θ value | 8.0589e-150 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.850814 (rank : 4) | θ value | 6.17048e-150 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
ARH_HUMAN
|
||||||
| NC score | 0.588098 (rank : 5) | θ value | 3.18553e-13 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SW96, Q6TQS9, Q8N2Y0, Q9UFI9 | Gene names | LDLRAP1, ARH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein). | |||||
|
ARH_MOUSE
|
||||||
| NC score | 0.576376 (rank : 6) | θ value | 1.33837e-11 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C142, Q6NWV6, Q8VDQ0 | Gene names | Ldlrap1, Arh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Low density lipoprotein receptor adapter protein 1 (Autosomal recessive hypercholesterolemia protein homolog). | |||||
|
DAB1_MOUSE
|
||||||
| NC score | 0.397747 (rank : 7) | θ value | 1.53129e-07 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97318, P97316, P97317 | Gene names | Dab1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.390019 (rank : 8) | θ value | 1.53129e-07 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
CAPON_HUMAN
|
||||||
| NC score | 0.351611 (rank : 9) | θ value | 0.000121331 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75052, O43564 | Gene names | NOS1AP, CAPON, KIAA0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
CAPON_MOUSE
|
||||||
| NC score | 0.340742 (rank : 10) | θ value | 0.00020696 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D3A8, Q80TZ6 | Gene names | Nos1ap, Capon, Kiaa0464 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase protein (C--terminal PDZ ligand of neuronal nitric oxide synthase protein) (Nitric oxide synthase 1 adaptor protein). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.337659 (rank : 11) | θ value | 5.44631e-05 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.334251 (rank : 12) | θ value | 4.1701e-05 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.213535 (rank : 13) | θ value | 0.00298849 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.203664 (rank : 14) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
FA43A_HUMAN
|
||||||
| NC score | 0.200841 (rank : 15) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N2R8, Q8IXP4, Q8WZ07 | Gene names | FAM43A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
FA43A_MOUSE
|
||||||
| NC score | 0.200742 (rank : 16) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BUP8, Q8R231 | Gene names | Fam43a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43A. | |||||
|
ANKS1_MOUSE
|
||||||
| NC score | 0.165522 (rank : 17) | θ value | 3.41135e-07 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59672, Q6ZQG0 | Gene names | Anks1a, Anks1, Kiaa0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A. | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.157028 (rank : 18) | θ value | 3.41135e-07 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.122400 (rank : 19) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
SHC2_MOUSE
|
||||||
| NC score | 0.110911 (rank : 20) | θ value | 0.00869519 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMC3 | Gene names | Shc2, Sck, ShcB | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 2 (SH2 domain protein C2) (Src homology 2 domain-containing-transforming protein C2) (Protein Sck) (Protein Sli). | |||||
|
SHC2_HUMAN
|
||||||
| NC score | 0.107710 (rank : 21) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98077, O60230, Q9NPL5, Q9UCX4 | Gene names | SHC2, SCK, SHCB | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 2 (SH2 domain protein C2) (Src homology 2 domain-containing-transforming protein C2) (Protein Sck). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.097242 (rank : 22) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.080909 (rank : 23) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.076326 (rank : 24) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.075060 (rank : 25) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.072236 (rank : 26) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
APBB1_MOUSE
|
||||||
| NC score | 0.071348 (rank : 27) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXJ1, O08642, Q3TPU0, Q8BNF4, Q8BSR9 | Gene names | Apbb1, Fe65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.066998 (rank : 28) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
FA43B_HUMAN
|
||||||
| NC score | 0.065991 (rank : 29) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZT52 | Gene names | FAM43B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM43B. | |||||
|
APBB1_HUMAN
|
||||||
| NC score | 0.065290 (rank : 30) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00213, Q96A93 | Gene names | APBB1, FE65 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1 (Fe65 protein). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.064535 (rank : 31) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.062510 (rank : 32) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.060421 (rank : 33) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.060343 (rank : 34) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.058907 (rank : 35) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.058027 (rank : 36) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.058004 (rank : 37) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.056768 (rank : 38) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.056732 (rank : 39) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.056324 (rank : 40) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.056206 (rank : 41) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.056161 (rank : 42) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.055693 (rank : 43) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.055298 (rank : 44) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.054294 (rank : 45) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.054284 (rank : 46) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.053253 (rank : 47) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.052779 (rank : 48) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.052453 (rank : 49) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
JIP2_MOUSE
|
||||||
| NC score | 0.052371 (rank : 50) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERE9, Q9CXI4 | Gene names | Mapk8ip2, Ib2, Jip2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.051346 (rank : 51) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.050498 (rank : 52) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.050309 (rank : 53) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.050163 (rank : 54) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.047602 (rank : 55) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.047536 (rank : 56) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.045956 (rank : 57) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.044443 (rank : 58) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.042647 (rank : 59) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.042296 (rank : 60) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.041291 (rank : 61) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.040992 (rank : 62) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
HNF1A_HUMAN
|
||||||
| NC score | 0.040799 (rank : 63) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.040625 (rank : 64) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.038732 (rank : 65) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.038691 (rank : 66) | θ value | 0.0563607 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.038505 (rank : 67) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.037596 (rank : 68) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.037201 (rank : 69) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.037140 (rank : 70) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.036783 (rank : 71) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.036569 (rank : 72) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.035188 (rank : 73) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.034974 (rank : 74) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.034950 (rank : 75) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.033956 (rank : 76) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.033425 (rank : 77) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.033238 (rank : 78) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.033116 (rank : 79) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.032783 (rank : 80) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
FBX43_HUMAN
|
||||||
| NC score | 0.032452 (rank : 81) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4G163 | Gene names | FBXO43, EMI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 43 (Endogenous meiotic inhibitor 2). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.028694 (rank : 82) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
TBCD4_MOUSE
|
||||||
| NC score | 0.028514 (rank : 83) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYJ6, Q5DU23, Q66JU2, Q6P2M2, Q8BMH6, Q8BXM2 | Gene names | Tbc1d4, Kiaa0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
TBCD4_HUMAN
|
||||||
| NC score | 0.027875 (rank : 84) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60343, Q5W0B9 | Gene names | TBC1D4, KIAA0603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 4 (Akt substrate of 160 kDa) (AS160). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.026831 (rank : 85) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.026092 (rank : 86) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.024513 (rank : 87) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.023392 (rank : 88) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
TBCD1_HUMAN
|
||||||
| NC score | 0.023155 (rank : 89) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86TI0, Q96K82, Q9UPP4 | Gene names | TBC1D1, KIAA1108 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.022350 (rank : 90) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.021794 (rank : 91) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.021271 (rank : 92) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.020473 (rank : 93) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PRKN2_HUMAN
|
||||||
| NC score | 0.019981 (rank : 94) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60260, Q5TFV8, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44, Q8WW07 | Gene names | PARK2, PRKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) (Parkinson juvenile disease protein 2) (Parkinson disease protein 2). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.019844 (rank : 95) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.019500 (rank : 96) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
DOK2_MOUSE
|
||||||
| NC score | 0.018975 (rank : 97) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.017783 (rank : 98) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.017509 (rank : 99) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
DTX2_MOUSE
|
||||||
| NC score | 0.016849 (rank : 100) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
GSLG1_MOUSE
|
||||||
| NC score | 0.016667 (rank : 101) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61543, Q9QZ40 | Gene names | Glg1, Esl1, Mg160, Selel | |||
|
Domain Architecture |
|
|||||
| Description | Golgi apparatus protein 1 precursor (Golgi sialoglycoprotein MG-160) (E-selectin ligand 1) (ESL-1) (Selel). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.015642 (rank : 102) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
TEAD1_HUMAN
|
||||||
| NC score | 0.015289 (rank : 103) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28347 | Gene names | TEAD1, TCF13, TEF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-1 (TEA domain family member 1) (TEAD-1) (Protein GT-IIC) (Transcription factor 13) (NTEF-1). | |||||
|
ADA29_HUMAN
|
||||||
| NC score | 0.014566 (rank : 104) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.014345 (rank : 105) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.013124 (rank : 106) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
TEAD1_MOUSE
|
||||||
| NC score | 0.013017 (rank : 107) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P30051 | Gene names | Tead1, Tcf13, Tef-1, Tef1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-1 (TEA domain family member 1) (TEAD-1) (Protein GT-IIC) (Transcription factor 13) (NTEF-1). | |||||
|
BPIL3_MOUSE
|
||||||
| NC score | 0.012956 (rank : 108) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BU51, Q80ZU8 | Gene names | Bpil3 | |||
|
Domain Architecture |

|
|||||
| Description | Bactericidal/permeability-increasing protein-like 3 precursor. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.012850 (rank : 109) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
B3A2_HUMAN
|
||||||
| NC score | 0.012133 (rank : 110) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04920, Q45EY5, Q969L3 | Gene names | SLC4A2, AE2, EPB3L1, HKB3, MPB3L | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 2 (Non-erythroid band 3-like protein) (AE2 anion exchanger) (Solute carrier family 4 member 2) (BND3L). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.011525 (rank : 111) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.011435 (rank : 112) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.011349 (rank : 113) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.011187 (rank : 114) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PXDC2_MOUSE
|
||||||
| NC score | 0.010826 (rank : 115) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.009585 (rank : 116) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.009267 (rank : 117) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.009226 (rank : 118) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
WWP1_MOUSE
|
||||||
| NC score | 0.008409 (rank : 119) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BZZ3, Q8BIV9, Q8VDP8 | Gene names | Wwp1 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP1 (EC 6.3.2.-) (WW domain- containing protein 1). | |||||
|
TLN2_HUMAN
|
||||||
| NC score | 0.007741 (rank : 120) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4G6 | Gene names | TLN2, KIAA0320 | |||
|
Domain Architecture |

|
|||||
| Description | Talin-2. | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.007224 (rank : 121) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
HXD12_HUMAN
|
||||||
| NC score | 0.006648 (rank : 122) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35452, Q9NS03 | Gene names | HOXD12, HOX4H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D12 (Hox-4H). | |||||
|
SN1L1_HUMAN
|
||||||
| NC score | 0.005383 (rank : 123) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57059, Q5R2V5, Q86YJ2 | Gene names | SNF1LK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK). | |||||
|
PVRL1_HUMAN
|
||||||
| NC score | 0.004926 (rank : 124) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15223, O75465, Q9HBE6, Q9HBW2 | Gene names | PVRL1, HVEC, PRR1 | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 1 precursor (Herpes virus entry mediator C) (HveC) (Nectin-1) (Herpesvirus Ig-like receptor) (HIgR) (CD111 antigen). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.004458 (rank : 125) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
GBX1_HUMAN
|
||||||
| NC score | 0.003320 (rank : 126) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14549 | Gene names | GBX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein GBX-1 (Gastrulation and brain-specific homeobox protein 1). | |||||