Please be patient as the page loads
|
PI4KB_HUMAN
|
||||||
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PI4KB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996567 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | 3.60269e-49 (rank : 3) | NC score | 0.856250 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | 3.50572e-28 (rank : 4) | NC score | 0.792846 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 5) | NC score | 0.791219 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 6) | NC score | 0.805089 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 7) | NC score | 0.803833 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3CA_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 8) | NC score | 0.776610 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 9) | NC score | 0.776189 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 10) | NC score | 0.666821 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 1.37858e-24 (rank : 11) | NC score | 0.762576 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 12) | NC score | 0.762984 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 13) | NC score | 0.682624 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 4.43474e-23 (rank : 14) | NC score | 0.685129 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 15) | NC score | 0.765674 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 16) | NC score | 0.765917 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 17) | NC score | 0.765129 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 18) | NC score | 0.765207 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 19) | NC score | 0.428972 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 20) | NC score | 0.428603 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 21) | NC score | 0.394248 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
ATM_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 22) | NC score | 0.367484 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 23) | NC score | 0.372818 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 24) | NC score | 0.376881 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 25) | NC score | 0.384119 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 26) | NC score | 0.376479 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.035509 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.281844 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.285072 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.031046 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.016617 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
RPP25_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.069486 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE3 | Gene names | Rpp25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease P protein subunit p25 (EC 3.1.26.5) (RNase P protein subunit p25). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.018057 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.016357 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RBP1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.020537 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
NUAK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.006974 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H093 | Gene names | NUAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1) (SNF1/AMP kinase-related kinase) (SNARK). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.021960 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
G3BP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.014272 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.015544 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.021271 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.017490 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.006139 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.013896 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SGK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.006752 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00141, Q5TCN3, Q9UN56 | Gene names | SGK, SGK1 | |||
|
Domain Architecture |
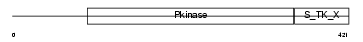
|
|||||
| Description | Serine/threonine-protein kinase Sgk1 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 1). | |||||
|
MARK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.005901 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
MCM3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.015203 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
NUAK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.006298 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
PRKY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.006355 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43930, O15348, O15349 | Gene names | PRKY | |||
|
Domain Architecture |
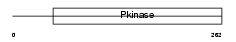
|
|||||
| Description | Serine/threonine-protein kinase PRKY (EC 2.7.11.1). | |||||
|
SGK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.006461 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVC6 | Gene names | Sgk, Sgk1 | |||
|
Domain Architecture |
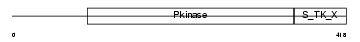
|
|||||
| Description | Serine/threonine-protein kinase Sgk1 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 1). | |||||
|
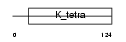
ATS9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.005178 (rank : 83) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2N4, Q9NR29 | Gene names | ADAMTS9, KIAA1312 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 9) (ADAM-TS 9) (ADAM-TS9). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.005086 (rank : 84) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
LAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.003449 (rank : 87) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
SIRT5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.025563 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2C6 | Gene names | Sirt5, Sir2l5 | |||
|
Domain Architecture |
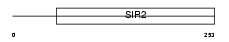
|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.052355 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.006365 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
PRKX_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.005791 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q922R0, Q3UCD1, Q8BHD6, Q9QZ12 | Gene names | Prkx, Pkare | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRKX (EC 2.7.11.1) (PKA-related protein kinase). | |||||
|
RSNL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.008840 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CI96, Q8BW09, Q921Q4, Q9D2S6 | Gene names | Rsnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin-like protein 2. | |||||
|
ZBTB6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | -0.001994 (rank : 89) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
IF16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.010485 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.009709 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KS6A4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.005378 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75676, O75585, Q53ES8 | Gene names | RPS6KA4, MSK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (Ribosomal protein kinase B) (RSKB). | |||||
|
KS6A4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.005302 (rank : 82) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2B9, Q91X18 | Gene names | Rps6ka4, Msk2 | |||
|
Domain Architecture |
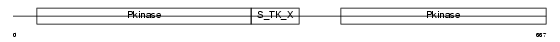
|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (RSK-like protein kinase) (RLSK). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.014960 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
NUMB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.017307 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
P3H3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.011668 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
PGFRB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.002617 (rank : 88) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P05622 | Gene names | Pdgfrb, Pdgfr | |||
|
Domain Architecture |

|
|||||
| Description | Beta platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-beta) (CD140b antigen). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007275 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.016317 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CP11A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.004606 (rank : 86) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ82 | Gene names | Cyp11a1, Cyp11a | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
GP143_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.010707 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51810 | Gene names | GPR143, OA1 | |||
|
Domain Architecture |
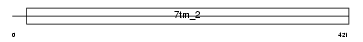
|
|||||
| Description | G-protein coupled receptor 143 (Ocular albinism type 1 protein). | |||||
|
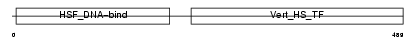
HSF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.012825 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
HSF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.012851 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38533, Q64157, Q8VEJ0, Q9Z2L8 | Gene names | Hsf2 | |||
|
Domain Architecture |
|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.005591 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.012650 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
NOL8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.007667 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.008208 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.007951 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PRKX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005019 (rank : 85) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51817 | Gene names | PRKX, PKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRKX (EC 2.7.11.1) (Protein kinase PKX1). | |||||
|
PRP40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.009000 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.058193 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.065060 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.058423 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RIMS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.066807 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.066816 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.050795 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.059359 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051180 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.071677 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.070747 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.996567 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.856250 (rank : 3) | θ value | 3.60269e-49 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.805089 (rank : 4) | θ value | 2.96777e-27 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.803833 (rank : 5) | θ value | 1.12775e-26 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.792846 (rank : 6) | θ value | 3.50572e-28 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.791219 (rank : 7) | θ value | 1.02001e-27 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.776610 (rank : 8) | θ value | 2.77775e-25 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.776189 (rank : 9) | θ value | 3.62785e-25 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.765917 (rank : 10) | θ value | 2.87452e-22 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.765674 (rank : 11) | θ value | 2.87452e-22 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.765207 (rank : 12) | θ value | 5.42112e-21 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.765129 (rank : 13) | θ value | 4.15078e-21 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.762984 (rank : 14) | θ value | 1.5242e-23 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.762576 (rank : 15) | θ value | 1.37858e-24 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.685129 (rank : 16) | θ value | 4.43474e-23 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.682624 (rank : 17) | θ value | 1.99067e-23 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.666821 (rank : 18) | θ value | 6.18819e-25 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.428972 (rank : 19) | θ value | 5.81887e-07 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.428603 (rank : 20) | θ value | 9.92553e-07 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.394248 (rank : 21) | θ value | 1.43324e-05 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.384119 (rank : 22) | θ value | 2.44474e-05 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.376881 (rank : 23) | θ value | 2.44474e-05 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.376479 (rank : 24) | θ value | 3.19293e-05 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.372818 (rank : 25) | θ value | 2.44474e-05 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.367484 (rank : 26) | θ value | 1.87187e-05 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.285072 (rank : 27) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.281844 (rank : 28) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.071677 (rank : 29) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.070747 (rank : 30) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
RPP25_MOUSE
|
||||||
| NC score | 0.069486 (rank : 31) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WE3 | Gene names | Rpp25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease P protein subunit p25 (EC 3.1.26.5) (RNase P protein subunit p25). | |||||
|
RIMS4_MOUSE
|
||||||
| NC score | 0.066816 (rank : 32) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| NC score | 0.066807 (rank : 33) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.065060 (rank : 34) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.059359 (rank : 35) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.058423 (rank : 36) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.058193 (rank : 37) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.052355 (rank : 38) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.051180 (rank : 39) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.050795 (rank : 40) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.035509 (rank : 41) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.031046 (rank : 42) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SIRT5_MOUSE
|
||||||
| NC score | 0.025563 (rank : 43) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2C6 | Gene names | Sirt5, Sir2l5 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.021960 (rank : 44) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.021271 (rank : 45) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
RBP1_MOUSE
|
||||||
| NC score | 0.020537 (rank : 46) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62172, Q9CRE5 | Gene names | Ralbp1, Rip1 | |||
|
Domain Architecture |

|
|||||
| Description | RalA-binding protein 1 (RalBP1) (Ral-interacting protein 1) (Dinitrophenyl S-glutathione ATPase) (DNP-SG ATPase). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.018057 (rank : 47) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.017490 (rank : 48) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
NUMB_MOUSE
|
||||||
| NC score | 0.017307 (rank : 49) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QZS3, P70422, Q8CIB1, Q9DC57, Q9QZR1, Q9QZS4 | Gene names | Numb | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (m-Numb) (m-Nb). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.016617 (rank : 50) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.016357 (rank : 51) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.016317 (rank : 52) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.015544 (rank : 53) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MCM3_MOUSE
|
||||||
| NC score | 0.015203 (rank : 54) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.014960 (rank : 55) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
G3BP_HUMAN
|
||||||
| NC score | 0.014272 (rank : 56) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.013896 (rank : 57) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
HSF2_MOUSE
|
||||||
| NC score | 0.012851 (rank : 58) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38533, Q64157, Q8VEJ0, Q9Z2L8 | Gene names | Hsf2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
HSF2_HUMAN
|
||||||
| NC score | 0.012825 (rank : 59) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.012650 (rank : 60) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
P3H3_HUMAN
|
||||||
| NC score | 0.011668 (rank : 61) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
GP143_HUMAN
|
||||||
| NC score | 0.010707 (rank : 62) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51810 | Gene names | GPR143, OA1 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 143 (Ocular albinism type 1 protein). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.010485 (rank : 63) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.009709 (rank : 64) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
PRP40_MOUSE
|
||||||
| NC score | 0.009000 (rank : 65) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R1C7, Q61049, Q8BQ76, Q8BRW4, Q8C2U1 | Gene names | Prpf40a, Fbp11, Fnbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 40 homolog A (Formin-binding protein 3) (Formin-binding protein 11) (FBP-11). | |||||
|
RSNL2_MOUSE
|
||||||
| NC score | 0.008840 (rank : 66) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CI96, Q8BW09, Q921Q4, Q9D2S6 | Gene names | Rsnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin-like protein 2. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.008208 (rank : 67) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.007951 (rank : 68) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
NOL8_MOUSE
|
||||||
| NC score | 0.007667 (rank : 69) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHX0, Q504M4, Q8CDJ7, Q9CUR0 | Gene names | Nol8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.007275 (rank : 70) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NUAK2_HUMAN
|
||||||
| NC score | 0.006974 (rank : 71) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H093 | Gene names | NUAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1) (SNF1/AMP kinase-related kinase) (SNARK). | |||||
|
SGK1_HUMAN
|
||||||
| NC score | 0.006752 (rank : 72) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00141, Q5TCN3, Q9UN56 | Gene names | SGK, SGK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Sgk1 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 1). | |||||
|
SGK1_MOUSE
|
||||||
| NC score | 0.006461 (rank : 73) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVC6 | Gene names | Sgk, Sgk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Sgk1 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 1). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.006365 (rank : 74) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
PRKY_HUMAN
|
||||||
| NC score | 0.006355 (rank : 75) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43930, O15348, O15349 | Gene names | PRKY | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRKY (EC 2.7.11.1). | |||||
|
NUAK2_MOUSE
|
||||||
| NC score | 0.006298 (rank : 76) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.006139 (rank : 77) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
MARK2_HUMAN
|
||||||
| NC score | 0.005901 (rank : 78) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7KZI7, Q15449, Q15524, Q5XGA3, Q68A18, Q96HB3, Q96RG0 | Gene names | MARK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MARK2 (EC 2.7.11.1) (MAP/microtubule affinity-regulating kinase 2) (ELKL motif kinase) (EMK1) (PAR1 homolog). | |||||
|
PRKX_MOUSE
|
||||||
| NC score | 0.005791 (rank : 79) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q922R0, Q3UCD1, Q8BHD6, Q9QZ12 | Gene names | Prkx, Pkare | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRKX (EC 2.7.11.1) (PKA-related protein kinase). | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.005591 (rank : 80) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KS6A4_HUMAN
|
||||||
| NC score | 0.005378 (rank : 81) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75676, O75585, Q53ES8 | Gene names | RPS6KA4, MSK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (Ribosomal protein kinase B) (RSKB). | |||||
|
KS6A4_MOUSE
|
||||||
| NC score | 0.005302 (rank : 82) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 866 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z2B9, Q91X18 | Gene names | Rps6ka4, Msk2 | |||
|
Domain Architecture |
|
|||||
| Description | Ribosomal protein S6 kinase alpha-4 (EC 2.7.11.1) (Nuclear mitogen- and stress-activated protein kinase 2) (90 kDa ribosomal protein S6 kinase 4) (RSK-like protein kinase) (RLSK). | |||||
|
ATS9_HUMAN
|
||||||
| NC score | 0.005178 (rank : 83) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2N4, Q9NR29 | Gene names | ADAMTS9, KIAA1312 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 9) (ADAM-TS 9) (ADAM-TS9). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.005086 (rank : 84) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
PRKX_HUMAN
|
||||||
| NC score | 0.005019 (rank : 85) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51817 | Gene names | PRKX, PKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRKX (EC 2.7.11.1) (Protein kinase PKX1). | |||||
|
CP11A_MOUSE
|
||||||
| NC score | 0.004606 (rank : 86) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ82 | Gene names | Cyp11a1, Cyp11a | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 11A1, mitochondrial precursor (EC 1.14.15.6) (CYPXIA1) (P450(scc)) (Cholesterol side-chain cleavage enzyme) (Cholesterol desmolase). | |||||
|
LAP2_MOUSE
|
||||||
| NC score | 0.003449 (rank : 87) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
PGFRB_MOUSE
|
||||||
| NC score | 0.002617 (rank : 88) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P05622 | Gene names | Pdgfrb, Pdgfr | |||
|
Domain Architecture |

|
|||||
| Description | Beta platelet-derived growth factor receptor precursor (EC 2.7.10.1) (PDGF-R-beta) (CD140b antigen). | |||||
|
ZBTB6_HUMAN
|
||||||
| NC score | -0.001994 (rank : 89) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||