Please be patient as the page loads
|
MCM3_MOUSE
|
||||||
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991129 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
MCM3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
MCM2_HUMAN
|
||||||
| θ value | 5.88354e-84 (rank : 3) | NC score | 0.938883 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49736, Q14577, Q15023, Q8N2V1, Q969W7, Q96AE1, Q9BRM7 | Gene names | MCM2, BM28, CDCL1, KIAA0030 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM7_HUMAN
|
||||||
| θ value | 1.71185e-83 (rank : 4) | NC score | 0.942788 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33993, Q15076, Q96D34, Q96GL1 | Gene names | MCM7, CDC47, MCM2 | |||
|
Domain Architecture |
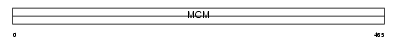
|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog) (P1.1-MCM3). | |||||
|
MCM2_MOUSE
|
||||||
| θ value | 2.23575e-83 (rank : 5) | NC score | 0.938702 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97310, O08971, O89057, Q8C2R0 | Gene names | Mcm2, Bm28, Cdcl1, Kiaa0030, Mcmd2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM6_MOUSE
|
||||||
| θ value | 1.44917e-82 (rank : 6) | NC score | 0.936989 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97311, Q80YQ7 | Gene names | Mcm6, Mcmd6, Mis5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (Mis5 homolog). | |||||
|
MCM7_MOUSE
|
||||||
| θ value | 2.47191e-82 (rank : 7) | NC score | 0.941927 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61881 | Gene names | Mcm7, Cdc47, Mcmd7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog). | |||||
|
MCM4_MOUSE
|
||||||
| θ value | 7.19216e-82 (rank : 8) | NC score | 0.932779 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM6_HUMAN
|
||||||
| θ value | 1.03855e-80 (rank : 9) | NC score | 0.936342 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14566, Q13504, Q99859 | Gene names | MCM6 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (p105MCM). | |||||
|
MCM5_HUMAN
|
||||||
| θ value | 4.82422e-78 (rank : 10) | NC score | 0.935627 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM5_MOUSE
|
||||||
| θ value | 6.96618e-77 (rank : 11) | NC score | 0.934811 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM4_HUMAN
|
||||||
| θ value | 1.45253e-74 (rank : 12) | NC score | 0.930872 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM8_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 13) | NC score | 0.912108 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MCM8_MOUSE
|
||||||
| θ value | 3.974e-56 (rank : 14) | NC score | 0.911917 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 15) | NC score | 0.064935 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 16) | NC score | 0.025335 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.058194 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.034840 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.025797 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.032381 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.032734 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.025051 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
VPS4A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.023723 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.025365 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CC45L_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.052124 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1X9, O70547, Q9QUK1, Q9R212 | Gene names | Cdc45l, Cdc45l2 | |||
|
Domain Architecture |

|
|||||
| Description | CDC45-related protein (PORC-PI-1). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.025188 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
STXB4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.029606 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.027133 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.025740 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
VPS4A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.020897 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.026836 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.015163 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CND3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.031567 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.036260 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.008361 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.042078 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.027356 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
OSBL8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.016110 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
STXB4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.029574 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.022911 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.025066 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CN145_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.022228 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.023598 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.022893 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.043324 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.033823 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RNF34_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.013818 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
SNUT1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.036975 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
STIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.010850 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.026935 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.019695 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
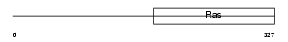
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.000986 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.028542 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.007914 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.015203 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.015114 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
RL6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.022540 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47911, Q6P5I2, Q925C3 | Gene names | Rpl6 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107). | |||||
|
SH24A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.026152 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.018487 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
ZIM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | -0.001883 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 718 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZV7 | Gene names | ZIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger imprinted 2. | |||||
|
CC45L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.035731 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75419, O60856, Q6UW54, Q9UP68 | Gene names | CDC45L, CDC45L2 | |||
|
Domain Architecture |

|
|||||
| Description | CDC45-related protein (PORC-PI-1) (Cdc45). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.010605 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.024007 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.023535 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
PAPS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.028971 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43252, O43841, O75332, Q96FB1, Q96TF4, Q9P1P9, Q9UE98 | Gene names | PAPSS1, ATPSK1, PAPSS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.028969 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60967 | Gene names | Papss1, Asapk, Atpsk1, Papss | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.029034 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95340, Q9BZL2, Q9P0G6, Q9UHM1, Q9UKD3, Q9UP30 | Gene names | PAPSS2, ATPSK2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 (PAPS synthetase 2) (PAPSS 2) (Sulfurylase kinase 2) (SK2) (SK 2) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.029043 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88428, Q9Z274 | Gene names | Papss2, Atpsk2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 (PAPS synthetase 2) (PAPSS 2) (Sulfurylase kinase 2) (SK2) (SK 2) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
CN155_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.017682 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.014148 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.023620 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
POT14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.006845 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
SNUT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.031101 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.013889 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.016422 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.000631 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
GP158_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.017038 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
KIF9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.002449 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV04, O35070 | Gene names | Kif9 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF9. | |||||
|
KU70_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.010419 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23475, O88212, Q62027, Q62382, Q62453 | Gene names | Xrcc6, G22p1, Ku70 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Ku autoantigen protein p70 homolog) (Ku70) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA- repair protein XRCC6). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.019774 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.016689 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
UBP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.003551 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
WDHD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.008763 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
APTX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.008176 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.022608 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.029001 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CDC37_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.014836 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |
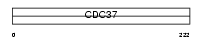
|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
DHB4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.007125 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
KPCD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | -0.001878 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15139 | Gene names | PRKD1, PKD, PKD1, PRKCM | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.012753 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.023403 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
PACN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.004509 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
RL6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.017202 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02878, Q2M3Q3, Q8WW97 | Gene names | RPL6 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107) (Neoplasm-related protein C140). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.019479 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.019162 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
MCM3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P25206, Q61492 | Gene names | Mcm3, Mcmd, Mcmd3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (P1-MCM3). | |||||
|
MCM3_HUMAN
|
||||||
| NC score | 0.991129 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P25205, Q92660, Q9BTR3, Q9NUE7 | Gene names | MCM3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM3 (DNA polymerase alpha holoenzyme-associated protein P1) (RLF subunit beta) (P102 protein) (P1-MCM3). | |||||
|
MCM7_HUMAN
|
||||||
| NC score | 0.942788 (rank : 3) | θ value | 1.71185e-83 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33993, Q15076, Q96D34, Q96GL1 | Gene names | MCM7, CDC47, MCM2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog) (P1.1-MCM3). | |||||
|
MCM7_MOUSE
|
||||||
| NC score | 0.941927 (rank : 4) | θ value | 2.47191e-82 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61881 | Gene names | Mcm7, Cdc47, Mcmd7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM7 (CDC47 homolog). | |||||
|
MCM2_HUMAN
|
||||||
| NC score | 0.938883 (rank : 5) | θ value | 5.88354e-84 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49736, Q14577, Q15023, Q8N2V1, Q969W7, Q96AE1, Q9BRM7 | Gene names | MCM2, BM28, CDCL1, KIAA0030 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM2_MOUSE
|
||||||
| NC score | 0.938702 (rank : 6) | θ value | 2.23575e-83 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P97310, O08971, O89057, Q8C2R0 | Gene names | Mcm2, Bm28, Cdcl1, Kiaa0030, Mcmd2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM2 (Minichromosome maintenance protein 2 homolog) (Nuclear protein BM28). | |||||
|
MCM6_MOUSE
|
||||||
| NC score | 0.936989 (rank : 7) | θ value | 1.44917e-82 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97311, Q80YQ7 | Gene names | Mcm6, Mcmd6, Mis5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (Mis5 homolog). | |||||
|
MCM6_HUMAN
|
||||||
| NC score | 0.936342 (rank : 8) | θ value | 1.03855e-80 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14566, Q13504, Q99859 | Gene names | MCM6 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM6 (p105MCM). | |||||
|
MCM5_HUMAN
|
||||||
| NC score | 0.935627 (rank : 9) | θ value | 4.82422e-78 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P33992, O60785, Q14578, Q9BTJ4, Q9BWL8 | Gene names | MCM5, CDC46 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM5_MOUSE
|
||||||
| NC score | 0.934811 (rank : 10) | θ value | 6.96618e-77 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
MCM4_MOUSE
|
||||||
| NC score | 0.932779 (rank : 11) | θ value | 7.19216e-82 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P49717, O89056 | Gene names | Mcm4, Cdc21, Mcmd4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM4_HUMAN
|
||||||
| NC score | 0.930872 (rank : 12) | θ value | 1.45253e-74 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
MCM8_HUMAN
|
||||||
| NC score | 0.912108 (rank : 13) | θ value | 1.36585e-56 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
MCM8_MOUSE
|
||||||
| NC score | 0.911917 (rank : 14) | θ value | 3.974e-56 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.064935 (rank : 15) | θ value | 3.19293e-05 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.058194 (rank : 16) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CC45L_MOUSE
|
||||||
| NC score | 0.052124 (rank : 17) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1X9, O70547, Q9QUK1, Q9R212 | Gene names | Cdc45l, Cdc45l2 | |||
|
Domain Architecture |

|
|||||
| Description | CDC45-related protein (PORC-PI-1). | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.043324 (rank : 18) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.042078 (rank : 19) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
SNUT1_HUMAN
|
||||||
| NC score | 0.036975 (rank : 20) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43290, Q53GB5 | Gene names | SART1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (U4/U6.U5 tri-snRNP-associated 110 kDa protein) (Squamous cell carcinoma antigen recognized by T cells 1) (SART-1) (hSART-1) (hSnu66). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.036260 (rank : 21) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
CC45L_HUMAN
|
||||||
| NC score | 0.035731 (rank : 22) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75419, O60856, Q6UW54, Q9UP68 | Gene names | CDC45L, CDC45L2 | |||
|
Domain Architecture |

|
|||||
| Description | CDC45-related protein (PORC-PI-1) (Cdc45). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.034840 (rank : 23) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.033823 (rank : 24) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.032734 (rank : 25) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.032381 (rank : 26) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CND3_HUMAN
|
||||||
| NC score | 0.031567 (rank : 27) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
SNUT1_MOUSE
|
||||||
| NC score | 0.031101 (rank : 28) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
STXB4_MOUSE
|
||||||
| NC score | 0.029606 (rank : 29) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 629 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WV89, Q5SUA6, Q5SUA8, Q5SUA9, Q8CFL1 | Gene names | Stxbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.029574 (rank : 30) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
PAPS2_MOUSE
|
||||||
| NC score | 0.029043 (rank : 31) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88428, Q9Z274 | Gene names | Papss2, Atpsk2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 (PAPS synthetase 2) (PAPSS 2) (Sulfurylase kinase 2) (SK2) (SK 2) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS2_HUMAN
|
||||||
| NC score | 0.029034 (rank : 32) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95340, Q9BZL2, Q9P0G6, Q9UHM1, Q9UKD3, Q9UP30 | Gene names | PAPSS2, ATPSK2 | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 2 (PAPS synthetase 2) (PAPSS 2) (Sulfurylase kinase 2) (SK2) (SK 2) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.029001 (rank : 33) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
PAPS1_HUMAN
|
||||||
| NC score | 0.028971 (rank : 34) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43252, O43841, O75332, Q96FB1, Q96TF4, Q9P1P9, Q9UE98 | Gene names | PAPSS1, ATPSK1, PAPSS | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
PAPS1_MOUSE
|
||||||
| NC score | 0.028969 (rank : 35) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60967 | Gene names | Papss1, Asapk, Atpsk1, Papss | |||
|
Domain Architecture |

|
|||||
| Description | Bifunctional 3'-phosphoadenosine 5'-phosphosulfate synthetase 1 (PAPS synthetase 1) (PAPSS 1) (Sulfurylase kinase 1) (SK1) (SK 1) [Includes: Sulfate adenylyltransferase (EC 2.7.7.4) (Sulfate adenylate transferase) (SAT) (ATP-sulfurylase); Adenylyl-sulfate kinase (EC 2.7.1.25) (Adenylylsulfate 3'-phosphotransferase) (APS kinase) (Adenosine-5'-phosphosulfate 3'-phosphotransferase) (3'- phosphoadenosine-5'-phosphosulfate synthetase)]. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.028542 (rank : 36) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.027356 (rank : 37) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.027133 (rank : 38) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.026935 (rank : 39) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.026836 (rank : 40) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.026152 (rank : 41) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.025797 (rank : 42) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.025740 (rank : 43) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.025365 (rank : 44) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.025335 (rank : 45) | θ value | 0.00298849 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.025188 (rank : 46) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.025066 (rank : 47) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.025051 (rank : 48) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.024007 (rank : 49) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
VPS4A_MOUSE
|
||||||
| NC score | 0.023723 (rank : 50) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEJ9, Q3TXT2 | Gene names | Vps4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A. | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.023620 (rank : 51) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.023598 (rank : 52) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.023535 (rank : 53) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.023403 (rank : 54) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.022911 (rank : 55) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.022893 (rank : 56) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.022608 (rank : 57) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
RL6_MOUSE
|
||||||
| NC score | 0.022540 (rank : 58) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P47911, Q6P5I2, Q925C3 | Gene names | Rpl6 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.022228 (rank : 59) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
VPS4A_HUMAN
|
||||||
| NC score | 0.020897 (rank : 60) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN37, Q8TF07, Q9UI03, Q9Y582 | Gene names | VPS4A, VPS4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vacuolar protein sorting-associating protein 4A (SKD2 protein) (hVPS4) (VPS4-1). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.019774 (rank : 61) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.019695 (rank : 62) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.019479 (rank : 63) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.019162 (rank : 64) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.018487 (rank : 65) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.017682 (rank : 66) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RL6_HUMAN
|
||||||
| NC score | 0.017202 (rank : 67) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02878, Q2M3Q3, Q8WW97 | Gene names | RPL6 | |||
|
Domain Architecture |

|
|||||
| Description | 60S ribosomal protein L6 (TAX-responsive enhancer element-binding protein 107) (TAXREB107) (Neoplasm-related protein C140). | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.017038 (rank : 68) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.016689 (rank : 69) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.016422 (rank : 70) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
OSBL8_HUMAN
|
||||||
| NC score | 0.016110 (rank : 71) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BZF1, Q8WXP8, Q9P277 | Gene names | OSBPL8, KIAA1451, ORP8, OSBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 8 (OSBP-related protein 8) (ORP-8). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.015203 (rank : 72) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.015163 (rank : 73) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.015114 (rank : 74) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
CDC37_MOUSE
|
||||||
| NC score | 0.014836 (rank : 75) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61081, Q3TGP0 | Gene names | Cdc37 | |||
|
Domain Architecture |

|
|||||
| Description | Hsp90 co-chaperone Cdc37 (Hsp90 chaperone protein kinase-targeting subunit) (p50Cdc37). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.014148 (rank : 76) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.013889 (rank : 77) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
RNF34_MOUSE
|
||||||
| NC score | 0.013818 (rank : 78) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99KR6, Q3UV45 | Gene names | Rnf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 34 (RING finger protein RIFF) (Phafin-1). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.012753 (rank : 79) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
STIP1_HUMAN
|
||||||
| NC score | 0.010850 (rank : 80) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31948 | Gene names | STIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) (Transformation-sensitive protein IEF SSP 3521) (NY- REN-11 antigen). | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.010605 (rank : 81) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
KU70_MOUSE
|
||||||
| NC score | 0.010419 (rank : 82) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23475, O88212, Q62027, Q62382, Q62453 | Gene names | Xrcc6, G22p1, Ku70 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 1 (EC 3.6.1.-) (ATP-dependent DNA helicase II 70 kDa subunit) (Ku autoantigen protein p70 homolog) (Ku70) (CTC box-binding factor 75 kDa subunit) (CTCBF) (CTC75) (DNA- repair protein XRCC6). | |||||
|
WDHD1_HUMAN
|
||||||
| NC score | 0.008763 (rank : 83) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75717 | Gene names | WDHD1, AND1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat and HMG-box DNA-binding protein 1 (Acidic nucleoplasmic DNA- binding protein 1) (And-1). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.008361 (rank : 84) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
APTX_HUMAN
|
||||||
| NC score | 0.008176 (rank : 85) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.007914 (rank : 86) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
DHB4_HUMAN
|
||||||
| NC score | 0.007125 (rank : 87) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51659 | Gene names | HSD17B4, EDH17B4 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal multifunctional enzyme type 2 (MFE-2) (D-bifunctional protein) (DBP) (17-beta-hydroxysteroid dehydrogenase 4) (17-beta-HSD 4) (D-3-hydroxyacyl-CoA dehydratase) (EC 4.2.1.107) (3-alpha,7- alpha,12-alpha-trihydroxy-5-beta-cholest-24-enoyl-CoA hydratase) (3- hydroxyacyl-CoA dehydrogenase) (EC 1.1.1.35). | |||||
|
POT14_HUMAN
|
||||||
| NC score | 0.006845 (rank : 88) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6S5H5, Q6S5H6, Q6S8J2 | Gene names | POTE14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 14. | |||||
|
PACN3_HUMAN
|
||||||
| NC score | 0.004509 (rank : 89) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKS6, Q9H331, Q9NWV9 | Gene names | PACSIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 3 (SH3 domain-containing protein 6511) (Endophilin I). | |||||
|
UBP4_HUMAN
|
||||||
| NC score | 0.003551 (rank : 90) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13107, O43452, O43453 | Gene names | USP4, UNP, UNPH | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 4 (EC 3.1.2.15) (Ubiquitin thioesterase 4) (Ubiquitin-specific-processing protease 4) (Deubiquitinating enzyme 4) (Ubiquitous nuclear protein homolog). | |||||
|
KIF9_MOUSE
|
||||||
| NC score | 0.002449 (rank : 91) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WV04, O35070 | Gene names | Kif9 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF9. | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.000986 (rank : 92) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.000631 (rank : 93) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
KPCD1_HUMAN
|
||||||
| NC score | -0.001878 (rank : 94) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15139 | Gene names | PRKD1, PKD, PKD1, PRKCM | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase D1 (EC 2.7.11.13) (nPKC-D1) (Protein kinase D) (Protein kinase C mu type) (nPKC-mu). | |||||
|
ZIM2_HUMAN
|
||||||
| NC score | -0.001883 (rank : 95) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 718 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZV7 | Gene names | ZIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger imprinted 2. | |||||