Please be patient as the page loads
|
PRKDC_HUMAN
|
||||||
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PRKDC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991668 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 3) | NC score | 0.693285 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 1.20211e-36 (rank : 4) | NC score | 0.687440 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 5) | NC score | 0.688724 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 2.26708e-35 (rank : 6) | NC score | 0.688829 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
ATM_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 7) | NC score | 0.655754 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 8) | NC score | 0.660001 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 9) | NC score | 0.613221 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 10) | NC score | 0.617804 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 11) | NC score | 0.418417 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 12) | NC score | 0.425972 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 13) | NC score | 0.424664 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 14) | NC score | 0.423641 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 15) | NC score | 0.425282 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 16) | NC score | 0.422847 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 17) | NC score | 0.424391 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 18) | NC score | 0.422747 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 19) | NC score | 0.448473 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 20) | NC score | 0.449485 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 21) | NC score | 0.384119 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 22) | NC score | 0.382334 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PK3CA_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 23) | NC score | 0.361526 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 24) | NC score | 0.361361 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 25) | NC score | 0.299951 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 26) | NC score | 0.338660 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 27) | NC score | 0.285459 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.333730 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 29) | NC score | 0.296407 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
XPO4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 30) | NC score | 0.070797 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0E2, Q5VUZ5, Q9H934 | Gene names | XPO4, KIAA1721 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-4 (Exp4). | |||||
|
XPO4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 31) | NC score | 0.070886 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESJ0, Q6ZPJ4 | Gene names | Xpo4, Kiaa1721 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-4 (Exp4). | |||||
|
CENPI_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.050882 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92674, Q5JWZ9, Q96ED0 | Gene names | CENPI, FSHPRH1, ICEN19, LRPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein I (CENP-I) (Interphase centromere complex protein 19) (Follicle-stimulating hormone primary response protein) (FSH primary response protein 1) (Leucine-rich primary response protein 1). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.015055 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
FZD6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.011381 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61089 | Gene names | Fzd6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (mFz6). | |||||
|
CX4NB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.029066 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70378 | Gene names | Cox4nb, Cox4al, Noc4 | |||
|
Domain Architecture |
|
|||||
| Description | Neighbor of COX4. | |||||
|
MYO1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.009817 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46735, P70244 | Gene names | Myo1b | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MIH-L). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.307833 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
SCYL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.015531 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
ACSL1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.013541 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33121, P41215, Q8N8V7, Q8TA99 | Gene names | ACSL1, FACL1, FACL2, LACS, LACS1, LACS2 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 1 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 1) (LACS 1) (Palmitoyl-CoA ligase 1) (Long-chain fatty acid CoA ligase 2) (Long-chain acyl-CoA synthetase 2) (LACS 2) (Acyl-CoA synthetase 1) (ACS1) (Palmitoyl-CoA ligase 2). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.014443 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
CETP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.061673 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11597, Q13987, Q13988 | Gene names | CETP | |||
|
Domain Architecture |
|
|||||
| Description | Cholesteryl ester transfer protein precursor (Lipid transfer protein I). | |||||
|
DMXL2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.017818 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
K0423_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.045830 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
E2F3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.021059 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.021131 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
RELN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.021333 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.017256 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
SMC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.014501 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
RHPN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.011784 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
SCYL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.012547 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 632 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3W7, Q96EF4, Q96ST4, Q9H7V5, Q9NVH3, Q9P2I7 | Gene names | SCYL2, CVAK104, KIAA1360 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
CREM_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.012928 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.004364 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
DDX54_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.005503 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
MAGB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.005236 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
PFTB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.013096 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2I1 | Gene names | Fntb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein farnesyltransferase subunit beta (EC 2.5.1.58) (CAAX farnesyltransferase subunit beta) (RAS proteins prenyltransferase beta) (FTase-beta). | |||||
|
UN93B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.014107 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCW4, O89077, Q710D2, Q8CIK1, Q8R3D0 | Gene names | Unc93b1, Unc93b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC93 homolog B1 (UNC-93B protein). | |||||
|
UNG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.022955 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |
|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.003463 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CUL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.009643 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.009648 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.005050 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.002503 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
FAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.011079 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.012294 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
FBX41_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.012823 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
GBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.007673 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.003195 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
RBBP8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.012731 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
RELN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.017044 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.010058 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
RIT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.001872 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92963, O00646, O00720 | Gene names | RIT1, RIBB, RIT, ROC1 | |||
|
Domain Architecture |
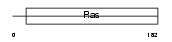
|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
RIT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.001866 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70426 | Gene names | Rit1, Rit | |||
|
Domain Architecture |
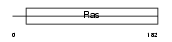
|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.003219 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | -0.002720 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
TRI32_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.008899 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
TRI32_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.008937 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
TTC12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.008002 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
UB2L3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.007520 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68036, P51966, P70653, Q9HAV1 | Gene names | UBE2L3, UBCE7, UBCH7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcH7) (E2-F1) (L-UBC). | |||||
|
UB2L3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.007520 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68037, P51966, P70653, Q9HAV1 | Gene names | Ube2l3, Ubce7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcM4). | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.006049 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
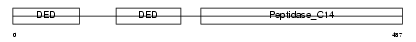
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CLH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.007181 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
CU056_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.011578 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.012894 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
ENSA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.011439 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43768, Q9NRZ0 | Gene names | ENSA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-endosulfine (ARPP-19e). | |||||
|
LRBA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.006354 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
NMNA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.008788 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BNJ3 | Gene names | Nmnat2, Kiaa0479 | |||
|
Domain Architecture |

|
|||||
| Description | Nicotinamide mononucleotide adenylyltransferase 2 (EC 2.7.7.1) (NMN adenylyltransferase 2). | |||||
|
PLD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.007547 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z280, O35911 | Gene names | Pld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (mPLD1). | |||||
|
PRPF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.014101 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |
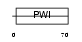
|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
PRPF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.013964 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
TEP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.003810 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97499 | Gene names | Tep1, Tp1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 homolog). | |||||
|
UTP20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.010056 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
ZDHC5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.006397 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |
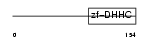
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ZDHC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.006374 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |
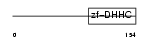
|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.991668 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.693285 (rank : 3) | θ value | 9.83387e-39 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.688829 (rank : 4) | θ value | 2.26708e-35 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.688724 (rank : 5) | θ value | 5.96599e-36 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.687440 (rank : 6) | θ value | 1.20211e-36 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.660001 (rank : 7) | θ value | 4.42448e-31 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.655754 (rank : 8) | θ value | 2.59387e-31 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.617804 (rank : 9) | θ value | 1.73987e-27 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.613221 (rank : 10) | θ value | 1.33217e-27 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.449485 (rank : 11) | θ value | 6.43352e-06 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.448473 (rank : 12) | θ value | 6.43352e-06 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.425972 (rank : 13) | θ value | 7.09661e-13 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.425282 (rank : 14) | θ value | 2.0648e-12 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.424664 (rank : 15) | θ value | 9.26847e-13 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.424391 (rank : 16) | θ value | 1.133e-10 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.423641 (rank : 17) | θ value | 1.58096e-12 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.422847 (rank : 18) | θ value | 2.69671e-12 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.422747 (rank : 19) | θ value | 2.52405e-10 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.418417 (rank : 20) | θ value | 5.43371e-13 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.384119 (rank : 21) | θ value | 2.44474e-05 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.382334 (rank : 22) | θ value | 2.44474e-05 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.361526 (rank : 23) | θ value | 0.00020696 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.361361 (rank : 24) | θ value | 0.00020696 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.338660 (rank : 25) | θ value | 0.000786445 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.333730 (rank : 26) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.307833 (rank : 27) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.299951 (rank : 28) | θ value | 0.000786445 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.296407 (rank : 29) | θ value | 0.00228821 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.285459 (rank : 30) | θ value | 0.00134147 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
XPO4_MOUSE
|
||||||
| NC score | 0.070886 (rank : 31) | θ value | 0.0961366 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESJ0, Q6ZPJ4 | Gene names | Xpo4, Kiaa1721 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-4 (Exp4). | |||||
|
XPO4_HUMAN
|
||||||
| NC score | 0.070797 (rank : 32) | θ value | 0.0961366 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C0E2, Q5VUZ5, Q9H934 | Gene names | XPO4, KIAA1721 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exportin-4 (Exp4). | |||||
|
CETP_HUMAN
|
||||||
| NC score | 0.061673 (rank : 33) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11597, Q13987, Q13988 | Gene names | CETP | |||
|
Domain Architecture |

|
|||||
| Description | Cholesteryl ester transfer protein precursor (Lipid transfer protein I). | |||||
|
CENPI_HUMAN
|
||||||
| NC score | 0.050882 (rank : 34) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92674, Q5JWZ9, Q96ED0 | Gene names | CENPI, FSHPRH1, ICEN19, LRPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein I (CENP-I) (Interphase centromere complex protein 19) (Follicle-stimulating hormone primary response protein) (FSH primary response protein 1) (Leucine-rich primary response protein 1). | |||||
|
K0423_MOUSE
|
||||||
| NC score | 0.045830 (rank : 35) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6A070 | Gene names | Kiaa0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
CX4NB_MOUSE
|
||||||
| NC score | 0.029066 (rank : 36) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70378 | Gene names | Cox4nb, Cox4al, Noc4 | |||
|
Domain Architecture |

|
|||||
| Description | Neighbor of COX4. | |||||
|
UNG2_HUMAN
|
||||||
| NC score | 0.022955 (rank : 37) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22674 | Gene names | UNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Uracil-DNA glycosylase 2 (EC 3.2.2.-) (UDG 2). | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.021333 (rank : 38) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
E2F3_MOUSE
|
||||||
| NC score | 0.021131 (rank : 39) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F3_HUMAN
|
||||||
| NC score | 0.021059 (rank : 40) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
DMXL2_HUMAN
|
||||||
| NC score | 0.017818 (rank : 41) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TDJ6, O94938 | Gene names | DMXL2, KIAA0856 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 2 (Rabconnectin-3). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.017256 (rank : 42) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
RELN_MOUSE
|
||||||
| NC score | 0.017044 (rank : 43) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
SCYL2_MOUSE
|
||||||
| NC score | 0.015531 (rank : 44) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.015055 (rank : 45) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.014501 (rank : 46) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.014443 (rank : 47) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
UN93B_MOUSE
|
||||||
| NC score | 0.014107 (rank : 48) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCW4, O89077, Q710D2, Q8CIK1, Q8R3D0 | Gene names | Unc93b1, Unc93b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC93 homolog B1 (UNC-93B protein). | |||||
|
PRPF3_HUMAN
|
||||||
| NC score | 0.014101 (rank : 49) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
PRPF3_MOUSE
|
||||||
| NC score | 0.013964 (rank : 50) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
ACSL1_HUMAN
|
||||||
| NC score | 0.013541 (rank : 51) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33121, P41215, Q8N8V7, Q8TA99 | Gene names | ACSL1, FACL1, FACL2, LACS, LACS1, LACS2 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 1 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 1) (LACS 1) (Palmitoyl-CoA ligase 1) (Long-chain fatty acid CoA ligase 2) (Long-chain acyl-CoA synthetase 2) (LACS 2) (Acyl-CoA synthetase 1) (ACS1) (Palmitoyl-CoA ligase 2). | |||||
|
PFTB_MOUSE
|
||||||
| NC score | 0.013096 (rank : 52) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2I1 | Gene names | Fntb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein farnesyltransferase subunit beta (EC 2.5.1.58) (CAAX farnesyltransferase subunit beta) (RAS proteins prenyltransferase beta) (FTase-beta). | |||||
|
CREM_MOUSE
|
||||||
| NC score | 0.012928 (rank : 53) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27699, P27698 | Gene names | Crem | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-responsive element modulator. | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.012894 (rank : 54) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
FBX41_MOUSE
|
||||||
| NC score | 0.012823 (rank : 55) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NS60, Q6P7W4, Q6ZPG1 | Gene names | Fbxo41, D6Ertd538e, Kiaa1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
RBBP8_HUMAN
|
||||||
| NC score | 0.012731 (rank : 56) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99708, O75371 | Gene names | RBBP8, CTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 8 (RBBP-8) (CtBP-interacting protein) (CtIP) (Retinoblastoma-interacting protein and myosin-like) (RIM). | |||||
|
SCYL2_HUMAN
|
||||||
| NC score | 0.012547 (rank : 57) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 632 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P3W7, Q96EF4, Q96ST4, Q9H7V5, Q9NVH3, Q9P2I7 | Gene names | SCYL2, CVAK104, KIAA1360 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.012294 (rank : 58) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
RHPN1_HUMAN
|
||||||
| NC score | 0.011784 (rank : 59) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
CU056_HUMAN
|
||||||
| NC score | 0.011578 (rank : 60) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A9, Q9NSE5 | Gene names | C21orf56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative protein C21orf56. | |||||
|
ENSA_HUMAN
|
||||||
| NC score | 0.011439 (rank : 61) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43768, Q9NRZ0 | Gene names | ENSA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-endosulfine (ARPP-19e). | |||||
|
FZD6_MOUSE
|
||||||
| NC score | 0.011381 (rank : 62) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61089 | Gene names | Fzd6 | |||
|
Domain Architecture |

|
|||||
| Description | Frizzled-6 precursor (Fz-6) (mFz6). | |||||
|
FAF1_MOUSE
|
||||||
| NC score | 0.011079 (rank : 63) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P54731 | Gene names | Faf1 | |||
|
Domain Architecture |

|
|||||
| Description | FAS-associated factor 1 (Protein FAF1). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.010058 (rank : 64) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
UTP20_HUMAN
|
||||||
| NC score | 0.010056 (rank : 65) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75691, Q9H3H4 | Gene names | UTP20, DRIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small subunit processome component 20 homolog (Down-regulated in metastasis protein) (Protein Key-1A6) (Novel nucleolar protein 73) (NNP73). | |||||
|
MYO1B_MOUSE
|
||||||
| NC score | 0.009817 (rank : 66) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46735, P70244 | Gene names | Myo1b | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ib (Myosin I alpha) (MMI-alpha) (MMIa) (MIH-L). | |||||
|
CUL3_MOUSE
|
||||||
| NC score | 0.009648 (rank : 67) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JLV5 | Gene names | Cul3 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
CUL3_HUMAN
|
||||||
| NC score | 0.009643 (rank : 68) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13618, O75415, Q569L3, Q9UBI8, Q9UET7 | Gene names | CUL3, KIAA0617 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-3 (CUL-3). | |||||
|
TRI32_MOUSE
|
||||||
| NC score | 0.008937 (rank : 69) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
TRI32_HUMAN
|
||||||
| NC score | 0.008899 (rank : 70) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
NMNA2_MOUSE
|
||||||
| NC score | 0.008788 (rank : 71) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BNJ3 | Gene names | Nmnat2, Kiaa0479 | |||
|
Domain Architecture |

|
|||||
| Description | Nicotinamide mononucleotide adenylyltransferase 2 (EC 2.7.7.1) (NMN adenylyltransferase 2). | |||||
|
TTC12_HUMAN
|
||||||
| NC score | 0.008002 (rank : 72) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H892, Q8N5H9, Q9NWY3 | Gene names | TTC12 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 12 (TPR repeat protein 12). | |||||
|
GBF1_HUMAN
|
||||||
| NC score | 0.007673 (rank : 73) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92538, Q5VXX3, Q96CK6, Q96HZ3, Q9H473 | Gene names | GBF1, KIAA0248 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (BFA-resistant GEF 1). | |||||
|
PLD1_MOUSE
|
||||||
| NC score | 0.007547 (rank : 74) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z280, O35911 | Gene names | Pld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline phosphatase 1) (Phosphatidylcholine-hydrolyzing phospholipase D1) (mPLD1). | |||||
|
UB2L3_HUMAN
|
||||||
| NC score | 0.007520 (rank : 75) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68036, P51966, P70653, Q9HAV1 | Gene names | UBE2L3, UBCE7, UBCH7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcH7) (E2-F1) (L-UBC). | |||||
|
UB2L3_MOUSE
|
||||||
| NC score | 0.007520 (rank : 76) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P68037, P51966, P70653, Q9HAV1 | Gene names | Ube2l3, Ubce7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 L3 (EC 6.3.2.19) (Ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) (UbcM4). | |||||
|
CLH2_HUMAN
|
||||||
| NC score | 0.007181 (rank : 77) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53675, Q14017, Q15808, Q15809 | Gene names | CLTCL1, CLH22, CLTCL, CLTD | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin heavy chain 2 (CLH-22). | |||||
|
ZDHC5_HUMAN
|
||||||
| NC score | 0.006397 (rank : 78) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0B5, Q6ZMF0, Q8TAK8, Q9H923, Q9UFI7 | Gene names | ZDHHC5, KIAA1748, ZNF375 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5) (Zinc finger protein 375). | |||||
|
ZDHC5_MOUSE
|
||||||
| NC score | 0.006374 (rank : 79) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDZ4, Q69ZB5, Q8R2X7 | Gene names | Zdhhc5, Kiaa1748 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC5 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 5) (DHHC-5). | |||||
|
LRBA_HUMAN
|
||||||
| NC score | 0.006354 (rank : 80) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50851, Q9H2U3, Q9H2U4 | Gene names | LRBA, BGL, CDC4L, LBA | |||
|
Domain Architecture |

|
|||||
| Description | Lipopolysaccharide-responsive and beige-like anchor protein (CDC4-like protein) (Beige-like protein). | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.006049 (rank : 81) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
DDX54_MOUSE
|
||||||
| NC score | 0.005503 (rank : 82) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K4L0 | Gene names | Ddx54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54). | |||||
|
MAGB2_HUMAN
|
||||||
| NC score | 0.005236 (rank : 83) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15479, O75860 | Gene names | MAGEB2 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen B2 (MAGE-B2 antigen) (DSS-AHC critical interval MAGE superfamily 6) (DAM6) (MAGE XP-2). | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.005050 (rank : 84) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.004364 (rank : 85) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
TEP1_MOUSE
|
||||||
| NC score | 0.003810 (rank : 86) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97499 | Gene names | Tep1, Tp1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 homolog). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.003463 (rank : 87) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.003219 (rank : 88) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.003195 (rank : 89) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.002503 (rank : 90) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
RIT1_HUMAN
|
||||||
| NC score | 0.001872 (rank : 91) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92963, O00646, O00720 | Gene names | RIT1, RIBB, RIT, ROC1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
RIT1_MOUSE
|
||||||
| NC score | 0.001866 (rank : 92) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70426 | Gene names | Rit1, Rit | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Rit1 (Ras-like protein expressed in many tissues) (Ras-like without CAAX protein 1). | |||||
|
SLK_HUMAN
|
||||||
| NC score | -0.002720 (rank : 93) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||