Please be patient as the page loads
|
CASP8_HUMAN
|
||||||
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
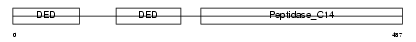
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CASP8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_MOUSE
|
||||||
| θ value | 1.3161e-184 (rank : 2) | NC score | 0.989068 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
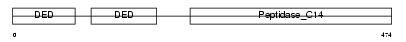
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 3) | NC score | 0.943414 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
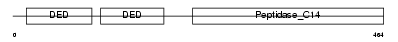
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP7_MOUSE
|
||||||
| θ value | 2.67181e-44 (rank : 4) | NC score | 0.907048 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| θ value | 1.98146e-39 (rank : 5) | NC score | 0.903133 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP3_HUMAN
|
||||||
| θ value | 9.83387e-39 (rank : 6) | NC score | 0.897227 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| θ value | 1.28434e-38 (rank : 7) | NC score | 0.897902 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP9_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 8) | NC score | 0.881557 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CFLAR_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 9) | NC score | 0.784602 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |
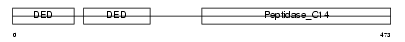
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP6_HUMAN
|
||||||
| θ value | 6.38894e-30 (rank : 10) | NC score | 0.880604 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| θ value | 1.4233e-29 (rank : 11) | NC score | 0.880512 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 12) | NC score | 0.832280 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
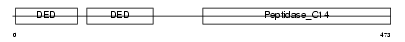
CFLAR_MOUSE
|
||||||
| θ value | 2.77775e-25 (rank : 13) | NC score | 0.750082 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 14) | NC score | 0.829102 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASPE_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 15) | NC score | 0.800064 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_MOUSE
|
||||||
| θ value | 1.52774e-15 (rank : 16) | NC score | 0.798897 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
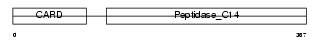
CASP4_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 17) | NC score | 0.634668 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP1_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 18) | NC score | 0.645743 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 19) | NC score | 0.629107 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP5_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 20) | NC score | 0.623138 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
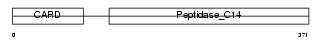
CASP4_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 21) | NC score | 0.595047 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
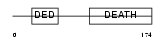
FADD_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 22) | NC score | 0.296855 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.273121 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
CASPC_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 24) | NC score | 0.589672 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
PEA15_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 25) | NC score | 0.199284 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
PEA15_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 26) | NC score | 0.198081 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.031613 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.032791 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.028939 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.026010 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.043238 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.043518 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.032201 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.016024 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.024702 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MKL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.026324 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.008439 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
DEDD_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.049328 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.015549 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
DEDD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.048133 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
GA2L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.016129 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.008699 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.022368 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.017836 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
DEDD2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.035978 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.023228 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.015036 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.021901 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.011921 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.019292 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
TRI39_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.005211 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESN2, Q8BPR5, Q8K0F7 | Gene names | Trim39, Rnf23, Tfp | |||
|
Domain Architecture |
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
AZI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.018333 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.005825 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
ZN622_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.012388 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969S3 | Gene names | ZNF622, ZPR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 622 (Zinc finger-like protein 9). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.007079 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
EP15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.014871 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.016892 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.008401 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.014294 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.017963 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.006049 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.012450 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
ICBR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.104955 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.989068 (rank : 2) | θ value | 1.3161e-184 (rank : 2) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.943414 (rank : 3) | θ value | 9.76566e-63 (rank : 3) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP7_MOUSE
|
||||||
| NC score | 0.907048 (rank : 4) | θ value | 2.67181e-44 (rank : 4) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| NC score | 0.903133 (rank : 5) | θ value | 1.98146e-39 (rank : 5) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP3_MOUSE
|
||||||
| NC score | 0.897902 (rank : 6) | θ value | 1.28434e-38 (rank : 7) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_HUMAN
|
||||||
| NC score | 0.897227 (rank : 7) | θ value | 9.83387e-39 (rank : 6) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.881557 (rank : 8) | θ value | 6.17384e-33 (rank : 8) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP6_HUMAN
|
||||||
| NC score | 0.880604 (rank : 9) | θ value | 6.38894e-30 (rank : 10) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| NC score | 0.880512 (rank : 10) | θ value | 1.4233e-29 (rank : 11) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_HUMAN
|
||||||
| NC score | 0.832280 (rank : 11) | θ value | 1.12775e-26 (rank : 12) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| NC score | 0.829102 (rank : 12) | θ value | 3.62785e-25 (rank : 14) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASPE_HUMAN
|
||||||
| NC score | 0.800064 (rank : 13) | θ value | 5.07402e-19 (rank : 15) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_MOUSE
|
||||||
| NC score | 0.798897 (rank : 14) | θ value | 1.52774e-15 (rank : 16) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CFLAR_HUMAN
|
||||||
| NC score | 0.784602 (rank : 15) | θ value | 1.0531e-32 (rank : 9) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.750082 (rank : 16) | θ value | 2.77775e-25 (rank : 13) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP1_MOUSE
|
||||||
| NC score | 0.645743 (rank : 17) | θ value | 4.59992e-12 (rank : 18) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP4_MOUSE
|
||||||
| NC score | 0.634668 (rank : 18) | θ value | 3.52202e-12 (rank : 17) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP1_HUMAN
|
||||||
| NC score | 0.629107 (rank : 19) | θ value | 1.9326e-10 (rank : 19) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP5_HUMAN
|
||||||
| NC score | 0.623138 (rank : 20) | θ value | 1.06045e-08 (rank : 20) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASP4_HUMAN
|
||||||
| NC score | 0.595047 (rank : 21) | θ value | 1.29631e-06 (rank : 21) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CASPC_MOUSE
|
||||||
| NC score | 0.589672 (rank : 22) | θ value | 0.000270298 (rank : 24) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
FADD_HUMAN
|
||||||
| NC score | 0.296855 (rank : 23) | θ value | 1.09739e-05 (rank : 22) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| NC score | 0.273121 (rank : 24) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_MOUSE
|
||||||
| NC score | 0.199284 (rank : 25) | θ value | 0.00175202 (rank : 25) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
PEA15_HUMAN
|
||||||
| NC score | 0.198081 (rank : 26) | θ value | 0.00228821 (rank : 26) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
ICBR_HUMAN
|
||||||
| NC score | 0.104955 (rank : 27) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
DEDD_MOUSE
|
||||||
| NC score | 0.049328 (rank : 28) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1L3, Q7TQH8, Q9R227 | Gene names | Dedd | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (DEDPro1). | |||||
|
DEDD_HUMAN
|
||||||
| NC score | 0.048133 (rank : 29) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75618, O60737 | Gene names | DEDD, DEDPRO1, DEFT | |||
|
Domain Architecture |

|
|||||
| Description | Death effector domain-containing protein (Death effector domain- containing testicular molecule) (DEDPro1) (FLDED-1). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.043518 (rank : 30) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.043238 (rank : 31) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
DEDD2_MOUSE
|
||||||
| NC score | 0.035978 (rank : 32) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.032791 (rank : 33) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.032201 (rank : 34) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.031613 (rank : 35) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.028939 (rank : 36) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MKL2_HUMAN
|
||||||
| NC score | 0.026324 (rank : 37) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULH7, Q68CT1, Q6UB16, Q86WW2, Q8N226 | Gene names | MKL2, KIAA1243, MRTFB | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B) (Megakaryoblastic leukemia 2). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.026010 (rank : 38) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.024702 (rank : 39) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.023228 (rank : 40) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.022368 (rank : 41) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.021901 (rank : 42) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.019292 (rank : 43) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
AZI1_MOUSE
|
||||||
| NC score | 0.018333 (rank : 44) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62036 | Gene names | Azi1, Az1, Azi | |||
|
Domain Architecture |

|
|||||
| Description | 5-azacytidine-induced protein 1 (Pre-acrosome localization protein 1). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.017963 (rank : 45) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.017836 (rank : 46) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.016892 (rank : 47) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GA2L1_MOUSE
|
||||||
| NC score | 0.016129 (rank : 48) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.016024 (rank : 49) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.015549 (rank : 50) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.015036 (rank : 51) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.014871 (rank : 52) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.014294 (rank : 53) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.012450 (rank : 54) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
ZN622_HUMAN
|
||||||
| NC score | 0.012388 (rank : 55) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969S3 | Gene names | ZNF622, ZPR9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 622 (Zinc finger-like protein 9). | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.011921 (rank : 56) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.008699 (rank : 57) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.008439 (rank : 58) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.008401 (rank : 59) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.007079 (rank : 60) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.006049 (rank : 61) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.005825 (rank : 62) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
TRI39_MOUSE
|
||||||
| NC score | 0.005211 (rank : 63) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 62 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESN2, Q8BPR5, Q8K0F7 | Gene names | Trim39, Rnf23, Tfp | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||