Please be patient as the page loads
|
CASP9_HUMAN
|
||||||
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CASP9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP3_HUMAN
|
||||||
| θ value | 1.08726e-37 (rank : 2) | NC score | 0.936279 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| θ value | 1.08726e-37 (rank : 3) | NC score | 0.936195 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP2_MOUSE
|
||||||
| θ value | 1.57e-36 (rank : 4) | NC score | 0.911443 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 5) | NC score | 0.909712 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 6) | NC score | 0.881557 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
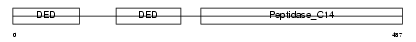
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_MOUSE
|
||||||
| θ value | 1.0531e-32 (rank : 7) | NC score | 0.888690 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
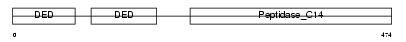
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP7_HUMAN
|
||||||
| θ value | 2.59387e-31 (rank : 8) | NC score | 0.920333 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_MOUSE
|
||||||
| θ value | 3.74554e-30 (rank : 9) | NC score | 0.921932 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| θ value | 1.12775e-26 (rank : 10) | NC score | 0.906067 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 11) | NC score | 0.904507 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 12) | NC score | 0.882521 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
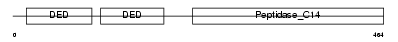
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASPE_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 13) | NC score | 0.879327 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASP1_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 14) | NC score | 0.768279 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 15) | NC score | 0.753705 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASPE_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 16) | NC score | 0.873384 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASP4_MOUSE
|
||||||
| θ value | 6.41864e-14 (rank : 17) | NC score | 0.753837 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |
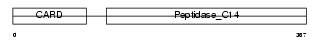
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP5_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 18) | NC score | 0.746895 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASPC_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 19) | NC score | 0.731565 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
CASP4_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 20) | NC score | 0.730560 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |
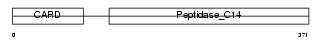
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CFLAR_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 21) | NC score | 0.585697 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
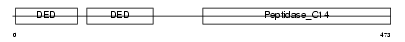
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 22) | NC score | 0.620466 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |
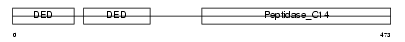
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
AMACR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.019904 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHK6, O43673, Q9Y3Q1 | Gene names | AMACR | |||
|
Domain Architecture |
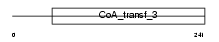
|
|||||
| Description | Alpha-methylacyl-CoA racemase (EC 5.1.99.4) (2-methylacyl-CoA racemase). | |||||
|
SPTA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.034090 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHX4, Q86WX5, Q8N9Y6 | Gene names | SPATA3, TSARG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1). | |||||
|
K0141_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.013534 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14154, Q969R4, Q96EU9 | Gene names | KIAA0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
P73L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.014752 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.014806 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
SFR16_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.012736 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
TGM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.005786 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLF6, Q8R0T9 | Gene names | Tgm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase K (EC 2.3.2.13) (Transglutaminase K) (TGase K) (TGK) (TG(K)) (Transglutaminase-1) (Epidermal TGase). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.006166 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
NKX22_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.001398 (rank : 38) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42586 | Gene names | Nkx2-2, Nkx-2.2, Nkx2b | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.2 (Homeobox protein NK-2 homolog B). | |||||
|
SAFB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.007837 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.005570 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
FADD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.118493 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |
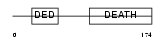
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.099931 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |
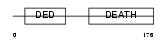
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
ICBR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.158569 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |
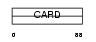
|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
PEA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.068624 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
PEA15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.069332 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP3_HUMAN
|
||||||
| NC score | 0.936279 (rank : 2) | θ value | 1.08726e-37 (rank : 2) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| NC score | 0.936195 (rank : 3) | θ value | 1.08726e-37 (rank : 3) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP7_MOUSE
|
||||||
| NC score | 0.921932 (rank : 4) | θ value | 3.74554e-30 (rank : 9) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| NC score | 0.920333 (rank : 5) | θ value | 2.59387e-31 (rank : 8) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP2_MOUSE
|
||||||
| NC score | 0.911443 (rank : 6) | θ value | 1.57e-36 (rank : 4) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_HUMAN
|
||||||
| NC score | 0.909712 (rank : 7) | θ value | 3.27365e-34 (rank : 5) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP6_MOUSE
|
||||||
| NC score | 0.906067 (rank : 8) | θ value | 1.12775e-26 (rank : 10) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_HUMAN
|
||||||
| NC score | 0.904507 (rank : 9) | θ value | 1.62847e-25 (rank : 11) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.888690 (rank : 10) | θ value | 1.0531e-32 (rank : 7) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.882521 (rank : 11) | θ value | 1.16704e-23 (rank : 12) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.881557 (rank : 12) | θ value | 6.17384e-33 (rank : 6) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPE_MOUSE
|
||||||
| NC score | 0.879327 (rank : 13) | θ value | 5.07402e-19 (rank : 13) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_HUMAN
|
||||||
| NC score | 0.873384 (rank : 14) | θ value | 1.38178e-16 (rank : 16) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASP1_MOUSE
|
||||||
| NC score | 0.768279 (rank : 15) | θ value | 4.29542e-18 (rank : 14) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP4_MOUSE
|
||||||
| NC score | 0.753837 (rank : 16) | θ value | 6.41864e-14 (rank : 17) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP1_HUMAN
|
||||||
| NC score | 0.753705 (rank : 17) | θ value | 1.63225e-17 (rank : 15) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP5_HUMAN
|
||||||
| NC score | 0.746895 (rank : 18) | θ value | 4.16044e-13 (rank : 18) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASPC_MOUSE
|
||||||
| NC score | 0.731565 (rank : 19) | θ value | 4.16044e-13 (rank : 19) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
CASP4_HUMAN
|
||||||
| NC score | 0.730560 (rank : 20) | θ value | 5.43371e-13 (rank : 20) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CFLAR_HUMAN
|
||||||
| NC score | 0.620466 (rank : 21) | θ value | 8.40245e-06 (rank : 22) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.585697 (rank : 22) | θ value | 4.92598e-06 (rank : 21) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
ICBR_HUMAN
|
||||||
| NC score | 0.158569 (rank : 23) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
FADD_HUMAN
|
||||||
| NC score | 0.118493 (rank : 24) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| NC score | 0.099931 (rank : 25) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_MOUSE
|
||||||
| NC score | 0.069332 (rank : 26) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
PEA15_HUMAN
|
||||||
| NC score | 0.068624 (rank : 27) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
SPTA3_HUMAN
|
||||||
| NC score | 0.034090 (rank : 28) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHX4, Q86WX5, Q8N9Y6 | Gene names | SPATA3, TSARG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 3 (Testis spermatocyte apoptosis- related protein 1) (Testis and spermatogenesis cell-related protein 1). | |||||
|
AMACR_HUMAN
|
||||||
| NC score | 0.019904 (rank : 29) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHK6, O43673, Q9Y3Q1 | Gene names | AMACR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-methylacyl-CoA racemase (EC 5.1.99.4) (2-methylacyl-CoA racemase). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.014806 (rank : 30) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.014752 (rank : 31) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
K0141_HUMAN
|
||||||
| NC score | 0.013534 (rank : 32) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14154, Q969R4, Q96EU9 | Gene names | KIAA0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
SFR16_MOUSE
|
||||||
| NC score | 0.012736 (rank : 33) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFC7, Q8CFC8, Q9Z2N3 | Gene names | Sfrs16, Clasp, Swap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 16 (Suppressor of white-apricot homolog 2) (Clk4-associating SR-related protein). | |||||
|
SAFB1_HUMAN
|
||||||
| NC score | 0.007837 (rank : 34) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15424, O60406, Q59HH8 | Gene names | SAFB, HAP, HET, SAFB1 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B (Scaffold attachment factor B1) (SAF-B) (HSP27 estrogen response element-TATA box-binding protein) (HSP27 ERE- TATA-binding protein). | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.006166 (rank : 35) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
TGM1_MOUSE
|
||||||
| NC score | 0.005786 (rank : 36) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JLF6, Q8R0T9 | Gene names | Tgm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein-glutamine gamma-glutamyltransferase K (EC 2.3.2.13) (Transglutaminase K) (TGase K) (TGK) (TG(K)) (Transglutaminase-1) (Epidermal TGase). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.005570 (rank : 37) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
NKX22_MOUSE
|
||||||
| NC score | 0.001398 (rank : 38) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 33 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42586 | Gene names | Nkx2-2, Nkx-2.2, Nkx2b | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.2 (Homeobox protein NK-2 homolog B). | |||||