Please be patient as the page loads
|
CASPA_HUMAN
|
||||||
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
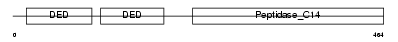
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CASPA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 9.76566e-63 (rank : 2) | NC score | 0.943414 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
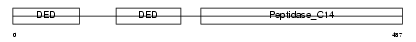
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_MOUSE
|
||||||
| θ value | 5.35859e-61 (rank : 3) | NC score | 0.946179 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
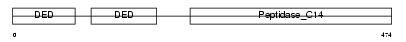
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP7_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 4) | NC score | 0.908356 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| θ value | 2.67802e-36 (rank : 5) | NC score | 0.905382 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP3_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 6) | NC score | 0.897133 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 7) | NC score | 0.895910 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP2_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 8) | NC score | 0.854187 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 9) | NC score | 0.854297 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP6_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 10) | NC score | 0.880837 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| θ value | 8.08199e-25 (rank : 11) | NC score | 0.879405 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP9_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 12) | NC score | 0.882521 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CFLAR_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 13) | NC score | 0.752506 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |
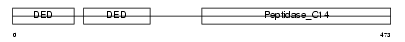
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP1_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 14) | NC score | 0.707534 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASPE_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 15) | NC score | 0.818249 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
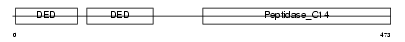
CFLAR_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 16) | NC score | 0.706891 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP1_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 17) | NC score | 0.688689 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP5_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 18) | NC score | 0.684824 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASPE_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 19) | NC score | 0.813933 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
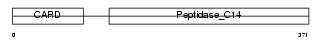
CASP4_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 20) | NC score | 0.655603 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
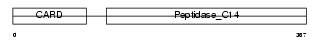
CASP4_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 21) | NC score | 0.678461 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASPC_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 22) | NC score | 0.651440 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
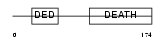
FADD_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 23) | NC score | 0.274936 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 24) | NC score | 0.246117 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.163259 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
PEA15_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 26) | NC score | 0.164036 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.015917 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NBN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.037218 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.015975 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.012576 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.021324 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.011759 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
HNRPC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.020265 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
PDE6B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.014557 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.013658 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
MED21_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.045842 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13503, Q92811 | Gene names | SURB7, MED21, SRB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 21 (RNA polymerase II holoenzyme component SRB7) (RNAPII complex component SRB7). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.012218 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
DEDD2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.022665 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
K0179_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.017427 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14684, Q8TBZ4 | Gene names | KIAA0179 | |||
|
Domain Architecture |
|
|||||
| Description | Protein KIAA0179. | |||||
|
CDK9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | -0.001757 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J95, Q3U002, Q3UMY2, Q3UPT3, Q3UQI6, Q8BTN0 | Gene names | Cdk9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division protein kinase 9 (EC 2.7.11.22) (EC 2.7.11.23) (Cyclin- dependent kinase 9). | |||||
|
COPG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.019631 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
MRCKA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.001106 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
NUP98_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.018408 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
PDE6B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.011747 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
PTK7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | -0.002133 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13308, Q13417 | Gene names | PTK7, CCK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase-like 7 precursor (Colon carcinoma kinase 4) (CCK-4). | |||||
|
CCD37_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.012389 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VN0, Q6AXD6 | Gene names | Ccdc37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
K0056_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.011828 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.008517 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
URP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.008500 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQL6, Q8IX34, Q8IYH2, Q9NWM2, Q9NXQ3 | Gene names | URP1, C20orf42, KIND1 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 1 (Kindlin-1) (Kindlerin) (Kindlin syndrome protein). | |||||
|
COPG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.017836 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
COPG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.018074 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.008765 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.007529 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
ICBR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.140770 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.946179 (rank : 2) | θ value | 5.35859e-61 (rank : 3) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.943414 (rank : 3) | θ value | 9.76566e-63 (rank : 2) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP7_MOUSE
|
||||||
| NC score | 0.908356 (rank : 4) | θ value | 5.39604e-37 (rank : 4) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| NC score | 0.905382 (rank : 5) | θ value | 2.67802e-36 (rank : 5) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP3_MOUSE
|
||||||
| NC score | 0.897133 (rank : 6) | θ value | 4.14116e-29 (rank : 6) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_HUMAN
|
||||||
| NC score | 0.895910 (rank : 7) | θ value | 1.73987e-27 (rank : 7) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.882521 (rank : 8) | θ value | 1.16704e-23 (rank : 12) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP6_HUMAN
|
||||||
| NC score | 0.880837 (rank : 9) | θ value | 2.12685e-25 (rank : 10) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| NC score | 0.879405 (rank : 10) | θ value | 8.08199e-25 (rank : 11) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_HUMAN
|
||||||
| NC score | 0.854297 (rank : 11) | θ value | 1.62847e-25 (rank : 9) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| NC score | 0.854187 (rank : 12) | θ value | 8.63488e-27 (rank : 8) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASPE_HUMAN
|
||||||
| NC score | 0.818249 (rank : 13) | θ value | 1.38178e-16 (rank : 15) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_MOUSE
|
||||||
| NC score | 0.813933 (rank : 14) | θ value | 1.86753e-13 (rank : 19) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CFLAR_HUMAN
|
||||||
| NC score | 0.752506 (rank : 15) | θ value | 1.99067e-23 (rank : 13) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP1_MOUSE
|
||||||
| NC score | 0.707534 (rank : 16) | θ value | 7.32683e-18 (rank : 14) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.706891 (rank : 17) | θ value | 1.16975e-15 (rank : 16) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CASP1_HUMAN
|
||||||
| NC score | 0.688689 (rank : 18) | θ value | 4.91457e-14 (rank : 17) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP5_HUMAN
|
||||||
| NC score | 0.684824 (rank : 19) | θ value | 1.09485e-13 (rank : 18) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASP4_MOUSE
|
||||||
| NC score | 0.678461 (rank : 20) | θ value | 4.0297e-08 (rank : 21) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP4_HUMAN
|
||||||
| NC score | 0.655603 (rank : 21) | θ value | 4.30538e-10 (rank : 20) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CASPC_MOUSE
|
||||||
| NC score | 0.651440 (rank : 22) | θ value | 4.0297e-08 (rank : 22) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
FADD_HUMAN
|
||||||
| NC score | 0.274936 (rank : 23) | θ value | 9.29e-05 (rank : 23) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| NC score | 0.246117 (rank : 24) | θ value | 0.00102713 (rank : 24) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
PEA15_MOUSE
|
||||||
| NC score | 0.164036 (rank : 25) | θ value | 0.0961366 (rank : 26) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
PEA15_HUMAN
|
||||||
| NC score | 0.163259 (rank : 26) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
ICBR_HUMAN
|
||||||
| NC score | 0.140770 (rank : 27) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57730 | Gene names | ICEBERG | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 inhibitor Iceberg. | |||||
|
MED21_HUMAN
|
||||||
| NC score | 0.045842 (rank : 28) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13503, Q92811 | Gene names | SURB7, MED21, SRB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of RNA polymerase II transcription subunit 21 (RNA polymerase II holoenzyme component SRB7) (RNAPII complex component SRB7). | |||||
|
NBN_MOUSE
|
||||||
| NC score | 0.037218 (rank : 29) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R207, O88981, Q3UY57, Q811I6, Q8CCY0, Q9R1X1 | Gene names | Nbn, Nbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nibrin (Nijmegen breakage syndrome protein 1 homolog) (Cell cycle regulatory protein p95). | |||||
|
DEDD2_MOUSE
|
||||||
| NC score | 0.022665 (rank : 30) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8QZV0, Q569Y9, Q8JZV1 | Gene names | Dedd2, Flame3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding death effector domain-containing protein 2 (FADD-like anti-apoptotic molecule 3) (DED-containing protein FLAME-3). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.021324 (rank : 31) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
HNRPC_HUMAN
|
||||||
| NC score | 0.020265 (rank : 32) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P07910, P22628, Q53EX2, Q59FD3, Q5FWE8, Q86SF8, Q86U45, Q96HK7, Q96HM4, Q96IY5, Q9BTS3 | Gene names | HNRPC | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
COPG2_HUMAN
|
||||||
| NC score | 0.019631 (rank : 33) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBF2 | Gene names | COPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
NUP98_HUMAN
|
||||||
| NC score | 0.018408 (rank : 34) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52948, Q8IUT2, Q8WYB0, Q96E54, Q9H3Q4, Q9NT02, Q9UF57, Q9UHX0, Q9Y6J4, Q9Y6J5 | Gene names | NUP98 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup98-Nup96 precursor [Contains: Nuclear pore complex protein Nup98 (Nucleoporin Nup98) (98 kDa nucleoporin); Nuclear pore complex protein Nup96 (Nucleoporin Nup96) (96 kDa nucleoporin)]. | |||||
|
COPG_HUMAN
|
||||||
| NC score | 0.018074 (rank : 35) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y678 | Gene names | COPG | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma (Gamma-coat protein) (Gamma-COP). | |||||
|
COPG2_MOUSE
|
||||||
| NC score | 0.017836 (rank : 36) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXK3 | Gene names | Copg2 | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit gamma-2 (Gamma-2 coat protein) (Gamma-2 COP). | |||||
|
K0179_HUMAN
|
||||||
| NC score | 0.017427 (rank : 37) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14684, Q8TBZ4 | Gene names | KIAA0179 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0179. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.015975 (rank : 38) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.015917 (rank : 39) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
PDE6B_MOUSE
|
||||||
| NC score | 0.014557 (rank : 40) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23440 | Gene names | Pde6b, Mpb, Pdeb, rd | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase subunit beta precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.013658 (rank : 41) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.012576 (rank : 42) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CCD37_MOUSE
|
||||||
| NC score | 0.012389 (rank : 43) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VN0, Q6AXD6 | Gene names | Ccdc37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.012218 (rank : 44) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.011828 (rank : 45) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.011759 (rank : 46) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
PDE6B_HUMAN
|
||||||
| NC score | 0.011747 (rank : 47) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35913, Q9BWH5 | Gene names | PDE6B, PDEB | |||
|
Domain Architecture |

|
|||||
| Description | Rod cGMP-specific 3',5'-cyclic phosphodiesterase beta-subunit precursor (EC 3.1.4.35) (GMP-PDE beta). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.008765 (rank : 48) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.008517 (rank : 49) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
URP1_HUMAN
|
||||||
| NC score | 0.008500 (rank : 50) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQL6, Q8IX34, Q8IYH2, Q9NWM2, Q9NXQ3 | Gene names | URP1, C20orf42, KIND1 | |||
|
Domain Architecture |

|
|||||
| Description | Unc-112-related protein 1 (Kindlin-1) (Kindlerin) (Kindlin syndrome protein). | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.007529 (rank : 51) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
MRCKA_HUMAN
|
||||||
| NC score | 0.001106 (rank : 52) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1999 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VT25, Q59GZ1, Q5H9N9, Q5T797, Q5VT26, Q5VT27, Q86XX2, Q86XX3, Q99646 | Gene names | CDC42BPA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK alpha (EC 2.7.11.1) (CDC42- binding protein kinase alpha) (Myotonic dystrophy kinase-related CDC42-binding kinase alpha) (Myotonic dystrophy protein kinase-like alpha) (MRCK alpha) (DMPK-like alpha). | |||||
|
CDK9_MOUSE
|
||||||
| NC score | -0.001757 (rank : 53) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99J95, Q3U002, Q3UMY2, Q3UPT3, Q3UQI6, Q8BTN0 | Gene names | Cdk9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division protein kinase 9 (EC 2.7.11.22) (EC 2.7.11.23) (Cyclin- dependent kinase 9). | |||||
|
PTK7_HUMAN
|
||||||
| NC score | -0.002133 (rank : 54) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 53 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13308, Q13417 | Gene names | PTK7, CCK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase-like 7 precursor (Colon carcinoma kinase 4) (CCK-4). | |||||