Please be patient as the page loads
|
FADD_HUMAN
|
||||||
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |
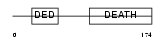
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FADD_HUMAN
|
||||||
| θ value | 1.18825e-76 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| θ value | 3.0428e-56 (rank : 2) | NC score | 0.961678 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |
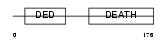
|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
CASP8_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 3) | NC score | 0.302427 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |
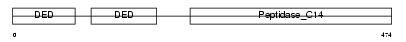
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
ANK1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 4) | NC score | 0.139090 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
CASP8_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 5) | NC score | 0.296855 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |
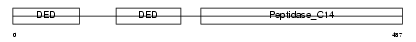
|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 6) | NC score | 0.140684 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
RIPK1_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 7) | NC score | 0.069830 (rank : 29) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 8) | NC score | 0.274936 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
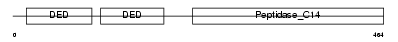
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
RIPK1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 9) | NC score | 0.067770 (rank : 32) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 10) | NC score | 0.126239 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CFLAR_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.252097 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |
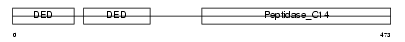
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.127528 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CRADD_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.100934 (rank : 23) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78560 | Gene names | CRADD, RAIDD | |||
|
Domain Architecture |
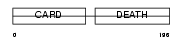
|
|||||
| Description | Death domain-containing protein CRADD (Caspase and RIP adapter with death domain) (RIP-associated protein with a death domain). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.037801 (rank : 67) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
CFLAR_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.234749 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |
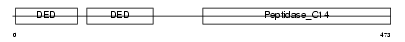
|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
TNR1A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.086142 (rank : 26) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TR10B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.069532 (rank : 30) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZM4, Q9JJL5, Q9JJL6 | Gene names | Tnfrsf10b, Dr5, Killer | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (MK). | |||||
|
TNR1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.083094 (rank : 27) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
CRADD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.080247 (rank : 28) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88843, Q9QUN0 | Gene names | Cradd, Raidd | |||
|
Domain Architecture |
|
|||||
| Description | Death domain-containing protein CRADD (Caspase and RIP adapter with death domain) (RIP-associated protein with a death domain). | |||||
|
CJ092_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.062997 (rank : 34) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.057749 (rank : 45) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.057441 (rank : 47) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANKS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.052356 (rank : 60) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.053604 (rank : 55) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
ANKY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.055091 (rank : 51) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
ANR16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.055812 (rank : 49) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
ANR22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.054523 (rank : 53) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ANR28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.057908 (rank : 43) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.057381 (rank : 48) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR29_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.050674 (rank : 62) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6D5, Q6ZWE8, Q96LU9 | Gene names | ANKRD29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 29. | |||||
|
ANR42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.050028 (rank : 66) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.053116 (rank : 57) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.057856 (rank : 44) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.060596 (rank : 39) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.058145 (rank : 41) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.057972 (rank : 42) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ASB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.050481 (rank : 63) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
ASB15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.052966 (rank : 58) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXK1 | Gene names | ASB15 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ASB15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.053671 (rank : 54) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ASB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.050442 (rank : 65) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.051754 (rank : 61) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
CASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.063762 (rank : 33) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.068437 (rank : 31) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.110157 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.108731 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.116610 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.117556 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.052570 (rank : 59) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
CASP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.062413 (rank : 35) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |
|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.062090 (rank : 36) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
CASP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.116410 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.116164 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.123354 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.125108 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.118493 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASPC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.050447 (rank : 64) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
CASPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.092452 (rank : 24) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.091083 (rank : 25) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CT086_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.053154 (rank : 56) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
INVS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.054845 (rank : 52) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
INVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.055605 (rank : 50) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
PEA15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.137249 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
PEA15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.136241 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.061850 (rank : 38) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.061887 (rank : 37) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.058600 (rank : 40) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.057651 (rank : 46) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
FADD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.18825e-76 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13158, Q14866 | Gene names | FADD, MORT1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
FADD_MOUSE
|
||||||
| NC score | 0.961678 (rank : 2) | θ value | 3.0428e-56 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61160, Q3TC37, Q61082 | Gene names | Fadd, Mort1 | |||
|
Domain Architecture |

|
|||||
| Description | FADD protein (FAS-associating death domain-containing protein) (Mediator of receptor induced toxicity). | |||||
|
CASP8_MOUSE
|
||||||
| NC score | 0.302427 (rank : 3) | θ value | 9.92553e-07 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O89110, O35669 | Gene names | Casp8 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASP8_HUMAN
|
||||||
| NC score | 0.296855 (rank : 4) | θ value | 1.09739e-05 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14790, O14676, Q14791, Q14792, Q14793, Q14794, Q14795, Q14796, Q15780, Q15806, Q53TT5, Q8TDI1, Q8TDI2, Q8TDI3, Q8TDI4, Q8TDI5, Q96T22, Q9C0K4, Q9UQ81 | Gene names | CASP8, MCH5 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-8 precursor (EC 3.4.22.-) (CASP-8) (ICE-like apoptotic protease 5) (MORT1-associated CED-3 homolog) (MACH) (FADD-homologous ICE/CED-3-like protease) (FADD-like ICE) (FLICE) (Apoptotic cysteine protease) (Apoptotic protease Mch-5) (CAP4) [Contains: Caspase-8 subunit p18; Caspase-8 subunit p10]. | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.274936 (rank : 5) | θ value | 9.29e-05 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CFLAR_HUMAN
|
||||||
| NC score | 0.252097 (rank : 6) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15519, O14673, O14674, O14675, O15137, O15138, O15356, O15510, O43618, O43619, O43620, O60458, O60459, Q96TE4, Q9UEW1 | Gene names | CFLAR, CASH, CLARP, MRIT | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
CFLAR_MOUSE
|
||||||
| NC score | 0.234749 (rank : 7) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35732, O35707, O35733 | Gene names | Cflar, Cash | |||
|
Domain Architecture |

|
|||||
| Description | CASP8 and FADD-like apoptosis regulator precursor (Cellular FLICE-like inhibitory protein) (c-FLIP) (Caspase-eight-related protein) (Casper) (Caspase-like apoptosis regulatory protein) (CLARP) (MACH-related inducer of toxicity) (MRIT) (Caspase homolog) (CASH) (Inhibitor of FLICE) (I-FLICE) (FADD-like antiapoptotic molecule 1) (FLAME-1) (Usurpin) [Contains: CASP8 and FADD-like apoptosis regulator subunit p43; CASP8 and FADD-like apoptosis regulator subunit p12]. | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.140684 (rank : 8) | θ value | 3.19293e-05 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.139090 (rank : 9) | θ value | 1.09739e-05 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
PEA15_HUMAN
|
||||||
| NC score | 0.137249 (rank : 10) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15121, O00511 | Gene names | PEA15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15 (Phosphoprotein enriched in diabetes) (PED). | |||||
|
PEA15_MOUSE
|
||||||
| NC score | 0.136241 (rank : 11) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q62048, Q3U5N7 | Gene names | Pea15 | |||
|
Domain Architecture |

|
|||||
| Description | Astrocytic phosphoprotein PEA-15. | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.127528 (rank : 12) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.126239 (rank : 13) | θ value | 0.00509761 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CASP7_MOUSE
|
||||||
| NC score | 0.125108 (rank : 14) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97864, O08669 | Gene names | Casp7, Lice2, Mch3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (LICE2 cysteine protease) (Apoptotic protease Mch-3) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP7_HUMAN
|
||||||
| NC score | 0.123354 (rank : 15) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55210, Q13364, Q96BA0 | Gene names | CASP7, MCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-7 precursor (EC 3.4.22.-) (CASP-7) (ICE-like apoptotic protease 3) (ICE-LAP3) (Apoptotic protease Mch-3) (CMH-1) [Contains: Caspase-7 subunit p20; Caspase-7 subunit p11]. | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.118493 (rank : 16) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
CASP3_MOUSE
|
||||||
| NC score | 0.117556 (rank : 17) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70677, O08668, Q8CHV5, Q9QWI4 | Gene names | Casp3, Cpp32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) (LICE) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP3_HUMAN
|
||||||
| NC score | 0.116610 (rank : 18) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42574, Q96AN1, Q96KP2 | Gene names | CASP3, CPP32 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-3 precursor (EC 3.4.22.-) (CASP-3) (Apopain) (Cysteine protease CPP32) (Yama protein) (CPP-32) (SREBP cleavage activity 1) (SCA-1) [Contains: Caspase-3 p17 subunit; Caspase-3 p12 subunit]. | |||||
|
CASP6_HUMAN
|
||||||
| NC score | 0.116410 (rank : 19) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55212, Q9BQE7 | Gene names | CASP6, MCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP6_MOUSE
|
||||||
| NC score | 0.116164 (rank : 20) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08738 | Gene names | Casp6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-6 precursor (EC 3.4.22.-) (CASP-6) (Apoptotic protease Mch-2) [Contains: Caspase-6 subunit p18; Caspase-6 subunit p11]. | |||||
|
CASP2_HUMAN
|
||||||
| NC score | 0.110157 (rank : 21) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42575, P42576, Q59F21, Q7KZL6, Q86UJ3, Q9BUP7, Q9BZK9 | Gene names | CASP2, ICH1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (ICH- 1L/1S) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CASP2_MOUSE
|
||||||
| NC score | 0.108731 (rank : 22) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29594, O08737, Q3TCM0, Q8C9H7, Q8K241 | Gene names | Casp2, Ich1, Nedd-2, Nedd2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-2 precursor (EC 3.4.22.-) (CASP-2) (ICH-1 protease) (NEDD2 protein) (Neural precursor cell expressed developmentally down- regulated protein 2) (NEDD-2) [Contains: Caspase-2 subunit p18; Caspase-2 subunit p13; Caspase-2 subunit p12]. | |||||
|
CRADD_HUMAN
|
||||||
| NC score | 0.100934 (rank : 23) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78560 | Gene names | CRADD, RAIDD | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-containing protein CRADD (Caspase and RIP adapter with death domain) (RIP-associated protein with a death domain). | |||||
|
CASPE_HUMAN
|
||||||
| NC score | 0.092452 (rank : 24) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P31944, O95823, Q3SYC9 | Gene names | CASP14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
CASPE_MOUSE
|
||||||
| NC score | 0.091083 (rank : 25) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89094 | Gene names | Casp14 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-14 precursor (EC 3.4.22.-) (CASP-14) (Mini-ICE) (MICE) [Contains: Caspase-14 subunit 1; Caspase-14 subunit 2]. | |||||
|
TNR1A_MOUSE
|
||||||
| NC score | 0.086142 (rank : 26) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR1A_HUMAN
|
||||||
| NC score | 0.083094 (rank : 27) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
CRADD_MOUSE
|
||||||
| NC score | 0.080247 (rank : 28) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88843, Q9QUN0 | Gene names | Cradd, Raidd | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-containing protein CRADD (Caspase and RIP adapter with death domain) (RIP-associated protein with a death domain). | |||||
|
RIPK1_MOUSE
|
||||||
| NC score | 0.069830 (rank : 29) | θ value | 4.1701e-05 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60855, Q3U0J3, Q8CD90 | Gene names | Ripk1, Rinp, Rip | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
TR10B_MOUSE
|
||||||
| NC score | 0.069532 (rank : 30) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZM4, Q9JJL5, Q9JJL6 | Gene names | Tnfrsf10b, Dr5, Killer | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (MK). | |||||
|
CASP1_MOUSE
|
||||||
| NC score | 0.068437 (rank : 31) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29452 | Gene names | Casp1, Il1bc | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
RIPK1_HUMAN
|
||||||
| NC score | 0.067770 (rank : 32) | θ value | 0.000121331 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 891 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13546, Q13180 | Gene names | RIPK1, RIP | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase RIP) (Cell death protein RIP) (Receptor-interacting protein). | |||||
|
CASP1_HUMAN
|
||||||
| NC score | 0.063762 (rank : 33) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29466, Q9UCN3 | Gene names | CASP1, IL1BC, IL1BCE | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-1 precursor (EC 3.4.22.36) (CASP-1) (Interleukin-1 beta convertase) (IL-1BC) (IL-1 beta-converting enzyme) (ICE) (Interleukin- 1 beta-converting enzyme) (p45) [Contains: Caspase-1 p20 subunit; Caspase-1 p10 subunit]. | |||||
|
CJ092_HUMAN
|
||||||
| NC score | 0.062997 (rank : 34) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
CASP4_MOUSE
|
||||||
| NC score | 0.062413 (rank : 35) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70343, O08735 | Gene names | Casp4, Casp11, Caspl, Ich3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (Caspase-11) (ICH-3 protease) [Contains: Caspase-4 subunit p10; Caspase-4 subunit p20]. | |||||
|
CASP5_HUMAN
|
||||||
| NC score | 0.062090 (rank : 36) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51878 | Gene names | CASP5, ICH3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-5 precursor (EC 3.4.22.-) (CASP-5) (ICH-3 protease) (TY protease) (ICE(rel)-III) [Contains: Caspase-5 subunit p20; Caspase-5 subunit p10]. | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.061887 (rank : 37) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.061850 (rank : 38) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.060596 (rank : 39) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.058600 (rank : 40) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.058145 (rank : 41) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.057972 (rank : 42) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.057908 (rank : 43) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.057856 (rank : 44) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.057749 (rank : 45) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.057651 (rank : 46) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.057441 (rank : 47) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.057381 (rank : 48) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR16_HUMAN
|
||||||
| NC score | 0.055812 (rank : 49) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.055605 (rank : 50) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
ANKY1_HUMAN
|
||||||
| NC score | 0.055091 (rank : 51) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2S6, Q8IYX5, Q8NDK5, Q9NX10 | Gene names | ANKMY1, TSAL1, ZMYND13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and MYND domain-containing protein 1 (Testis-specific ankyrin-like protein 1) (Zinc-finger MYND domain-containing protein 13). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.054845 (rank : 52) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
ANR22_HUMAN
|
||||||
| NC score | 0.054523 (rank : 53) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
ASB15_MOUSE
|
||||||
| NC score | 0.053671 (rank : 54) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.053604 (rank : 55) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
CT086_HUMAN
|
||||||
| NC score | 0.053154 (rank : 56) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZ19, Q4VXE6 | Gene names | C20orf86 | |||
|
Domain Architecture |

|
|||||
| Description | Putative uncharacterized protein C20orf86. | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.053116 (rank : 57) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ASB15_HUMAN
|
||||||
| NC score | 0.052966 (rank : 58) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXK1 | Gene names | ASB15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
CASP4_HUMAN
|
||||||
| NC score | 0.052570 (rank : 59) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49662 | Gene names | CASP4, ICH2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-4 precursor (EC 3.4.22.-) (CASP-4) (ICH-2 protease) (TX protease) (ICE(rel)-II) [Contains: Caspase-4 subunit 1; Caspase-4 subunit 2]. | |||||
|
ANKS6_HUMAN
|
||||||
| NC score | 0.052356 (rank : 60) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68DC2, Q5VSL0, Q5VSL2, Q5VSL3, Q5VSL4, Q68DB8, Q6P2R2, Q8N9L6, Q96D62 | Gene names | ANKS6, ANKRD14, SAMD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6) (Ankyrin repeat domain-containing protein 14). | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.051754 (rank : 61) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ANR29_HUMAN
|
||||||
| NC score | 0.050674 (rank : 62) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6D5, Q6ZWE8, Q96LU9 | Gene names | ANKRD29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 29. | |||||
|
ASB14_MOUSE
|
||||||
| NC score | 0.050481 (rank : 63) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
CASPC_MOUSE
|
||||||
| NC score | 0.050447 (rank : 64) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08736, Q3TT82 | Gene names | Casp12 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-12 precursor (EC 3.4.22.-) (CASP-12). | |||||
|
ASB3_HUMAN
|
||||||
| NC score | 0.050442 (rank : 65) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ANR42_HUMAN
|
||||||
| NC score | 0.050028 (rank : 66) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.037801 (rank : 67) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||