Please be patient as the page loads
|
P73L_MOUSE
|
||||||
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P73L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997089 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.964130 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
P53_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 4) | NC score | 0.907222 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
P53_MOUSE
|
||||||
| θ value | 6.7473e-72 (rank : 5) | NC score | 0.911548 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.085307 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
FOXI1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.034817 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q922I5, Q5SRI5, Q9D299 | Gene names | Foxi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I1. | |||||
|
EVI2B_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.118730 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
HELZ_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.056147 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.047570 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SAMD4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.072422 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU9 | Gene names | SAMD4, KIAA1053 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
ABI3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.051127 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.043775 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
CRX_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.021158 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54751 | Gene names | Crx | |||
|
Domain Architecture |
|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
DYR1B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.010222 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z188 | Gene names | Dyrk1b | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.050221 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
BRWD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.063835 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZH6, Q5VWA1 | Gene names | BRWD2, WDR11 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 2 (WD repeat protein 11). | |||||
|
EPHA4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.006846 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03137 | Gene names | Epha4, Sek, Sek1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (MPK-3). | |||||
|
FHOD1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.050492 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.045708 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.039167 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.034301 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
OXSR1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.005149 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95747, Q3LR53, Q7Z501, Q9UPQ1 | Gene names | OXSR1, KIAA1101, OSR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.035225 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
ZN313_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.025935 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ET26, Q3UFU8, Q8K5A2 | Gene names | Znf313, Zfp228, Zfp313, Znf228 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 313. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.046592 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.046796 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.034233 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DYM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.048429 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY3 | Gene names | Dym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.028065 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RSMB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.045130 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |
|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
DIAP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.033957 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.005450 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.037621 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
K0773_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.032341 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XD5, Q7L0D6, Q86T97 | Gene names | KIAA0773 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein KIAA0773. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.023946 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.025948 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.043750 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
RSMB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.039521 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |
|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.053108 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.016469 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.035163 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.031497 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
DYM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.040907 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTS9, Q6P2P5, Q8N2M0, Q9BVE9 | Gene names | DYM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin (Dyggve-Melchior-Clausen syndrome protein). | |||||
|
GGN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.031872 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.041389 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.029579 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NPL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.043547 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TAT6, Q8N3J1, Q9H8V2, Q9H964, Q9NWR5, Q9P229 | Gene names | NPLOC4, KIAA1499, NPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
NPL4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.043664 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60670 | Gene names | Nploc4, Kiaa1499, Npl4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.027905 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.022416 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SHC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.016187 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92529, Q8TAP2, Q9UCX5 | Gene names | SHC3, NSHC, SHCC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc) (Protein Rai). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.016561 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SOCS7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.023442 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.028767 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ABI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.023454 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62484, Q6PHU3 | Gene names | Abi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2). | |||||
|
CRX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.012746 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43186 | Gene names | CRX, CORD2 | |||
|
Domain Architecture |
|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.011113 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.026705 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EPHA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.004294 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54764 | Gene names | EPHA4, HEK8, SEK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (Receptor protein-tyrosine kinase HEK8). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.026440 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PRDM4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.000428 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||
|
PTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.015825 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.034989 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SF01_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.029124 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.025235 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.024956 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZIC5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.001973 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T25 | Gene names | ZIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 5 (Zinc finger protein of the cerebellum 5). | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.023819 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
CAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.016696 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |
|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CASP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.014806 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.021730 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
EVL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.017251 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.029997 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.023524 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ZN295_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.002769 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ3, Q6P4R0 | Gene names | ZNF295, KIAA1227, ZBTB21 | |||
|
Domain Architecture |
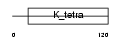
|
|||||
| Description | Zinc finger protein 295 (Zinc finger and BTB domain-containing protein 21). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.013511 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.023156 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
DIAP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.028992 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
F113A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.018737 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1Z5 | Gene names | Fam113a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A. | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.005243 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.034381 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
RED1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.007002 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.019996 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SETBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.021158 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SH2D3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.011017 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
SMAD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.015552 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMP1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.007353 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.997089 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.964130 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
P53_MOUSE
|
||||||
| NC score | 0.911548 (rank : 4) | θ value | 6.7473e-72 (rank : 5) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
|
P53_HUMAN
|
||||||
| NC score | 0.907222 (rank : 5) | θ value | 4.67263e-73 (rank : 4) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
EVI2B_MOUSE
|
||||||
| NC score | 0.118730 (rank : 6) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.085307 (rank : 7) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
SAMD4_HUMAN
|
||||||
| NC score | 0.072422 (rank : 8) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU9 | Gene names | SAMD4, KIAA1053 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 4. | |||||
|
BRWD2_HUMAN
|
||||||
| NC score | 0.063835 (rank : 9) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZH6, Q5VWA1 | Gene names | BRWD2, WDR11 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain and WD repeat domain-containing protein 2 (WD repeat protein 11). | |||||
|
HELZ_HUMAN
|
||||||
| NC score | 0.056147 (rank : 10) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42694 | Gene names | HELZ, KIAA0054 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable helicase with zinc-finger domain (EC 3.6.1.-). | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.053108 (rank : 11) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
ABI3_HUMAN
|
||||||
| NC score | 0.051127 (rank : 12) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2A4, Q9H0P6 | Gene names | ABI3, NESH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABI gene family member 3 (New molecule including SH3) (Nesh). | |||||
|
FHOD1_MOUSE
|
||||||
| NC score | 0.050492 (rank : 13) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P9Q4, Q8BMK2 | Gene names | Fhod1, Fhos1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.050221 (rank : 14) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
DYM_MOUSE
|
||||||
| NC score | 0.048429 (rank : 15) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHY3 | Gene names | Dym | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.047570 (rank : 16) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.046796 (rank : 17) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.046592 (rank : 18) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.045708 (rank : 19) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
RSMB_MOUSE
|
||||||
| NC score | 0.045130 (rank : 20) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.043775 (rank : 21) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.043750 (rank : 22) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
NPL4_MOUSE
|
||||||
| NC score | 0.043664 (rank : 23) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60670 | Gene names | Nploc4, Kiaa1499, Npl4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
NPL4_HUMAN
|
||||||
| NC score | 0.043547 (rank : 24) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TAT6, Q8N3J1, Q9H8V2, Q9H964, Q9NWR5, Q9P229 | Gene names | NPLOC4, KIAA1499, NPL4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear protein localization protein 4 homolog (Protein NPL4). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.041389 (rank : 25) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
DYM_HUMAN
|
||||||
| NC score | 0.040907 (rank : 26) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTS9, Q6P2P5, Q8N2M0, Q9BVE9 | Gene names | DYM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dymeclin (Dyggve-Melchior-Clausen syndrome protein). | |||||
|
RSMB_HUMAN
|
||||||
| NC score | 0.039521 (rank : 27) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.039167 (rank : 28) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.037621 (rank : 29) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.035225 (rank : 30) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.035163 (rank : 31) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.034989 (rank : 32) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
FOXI1_MOUSE
|
||||||
| NC score | 0.034817 (rank : 33) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q922I5, Q5SRI5, Q9D299 | Gene names | Foxi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I1. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.034381 (rank : 34) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.034301 (rank : 35) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.034233 (rank : 36) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
DIAP3_HUMAN
|
||||||
| NC score | 0.033957 (rank : 37) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
K0773_HUMAN
|
||||||
| NC score | 0.032341 (rank : 38) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XD5, Q7L0D6, Q86T97 | Gene names | KIAA0773 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein KIAA0773. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.031872 (rank : 39) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.031497 (rank : 40) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.029997 (rank : 41) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.029579 (rank : 42) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.029124 (rank : 43) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
DIAP3_MOUSE
|
||||||
| NC score | 0.028992 (rank : 44) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Z207 | Gene names | Diaph3, Diap3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3) (mDIA2) (p134mDIA2). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.028767 (rank : 45) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.028065 (rank : 46) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.027905 (rank : 47) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.026705 (rank : 48) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.026440 (rank : 49) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.025948 (rank : 50) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
ZN313_MOUSE
|
||||||
| NC score | 0.025935 (rank : 51) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ET26, Q3UFU8, Q8K5A2 | Gene names | Znf313, Zfp228, Zfp313, Znf228 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 313. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.025235 (rank : 52) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.024956 (rank : 53) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.023946 (rank : 54) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.023819 (rank : 55) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.023524 (rank : 56) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ABI2_MOUSE
|
||||||
| NC score | 0.023454 (rank : 57) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P62484, Q6PHU3 | Gene names | Abi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2). | |||||
|
SOCS7_MOUSE
|
||||||
| NC score | 0.023442 (rank : 58) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHQ2 | Gene names | Socs7, Cish7, Nap4 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 7 (SOCS-7). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.023156 (rank : 59) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.022416 (rank : 60) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.021730 (rank : 61) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
CRX_MOUSE
|
||||||
| NC score | 0.021158 (rank : 62) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54751 | Gene names | Crx | |||
|
Domain Architecture |

|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
SETBP_MOUSE
|
||||||
| NC score | 0.021158 (rank : 63) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z180, Q66JL8 | Gene names | Setbp1, Kiaa0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.019996 (rank : 64) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
F113A_MOUSE
|
||||||
| NC score | 0.018737 (rank : 65) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P1Z5 | Gene names | Fam113a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A. | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.017251 (rank : 66) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
CAP1_HUMAN
|
||||||
| NC score | 0.016696 (rank : 67) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.016561 (rank : 68) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.016469 (rank : 69) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
SHC3_HUMAN
|
||||||
| NC score | 0.016187 (rank : 70) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92529, Q8TAP2, Q9UCX5 | Gene names | SHC3, NSHC, SHCC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc) (Protein Rai). | |||||
|
PTC1_MOUSE
|
||||||
| NC score | 0.015825 (rank : 71) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.015552 (rank : 72) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
CASP9_HUMAN
|
||||||
| NC score | 0.014806 (rank : 73) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55211, O95348, Q92852, Q9BQ62, Q9UEQ3, Q9UIJ8 | Gene names | CASP9, MCH6 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-9 precursor (EC 3.4.22.-) (CASP-9) (ICE-like apoptotic protease 6) (ICE-LAP6) (Apoptotic protease Mch-6) (Apoptotic protease- activating factor 3) (APAF-3) [Contains: Caspase-9 subunit p35; Caspase-9 subunit p10]. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.013511 (rank : 74) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CRX_HUMAN
|
||||||
| NC score | 0.012746 (rank : 75) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43186 | Gene names | CRX, CORD2 | |||
|
Domain Architecture |

|
|||||
| Description | Cone-rod homeobox protein. | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.011113 (rank : 76) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
SH2D3_MOUSE
|
||||||
| NC score | 0.011017 (rank : 77) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZS8, Q9JME1 | Gene names | Sh2d3c, Chat, Shep1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (SH2 domain-containing Eph receptor- binding protein 1) (Cas/HEF1-associated signal transducer). | |||||
|
DYR1B_MOUSE
|
||||||
| NC score | 0.010222 (rank : 78) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z188 | Gene names | Dyrk1b | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1). | |||||
|
SMP1L_MOUSE
|
||||||
| NC score | 0.007353 (rank : 79) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TN29, Q3U798 | Gene names | Smap1l, Smap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1-like (Stromal membrane- associated protein 2). | |||||
|
RED1_HUMAN
|
||||||
| NC score | 0.007002 (rank : 80) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78563, O00395, O00465, O00691, O00692, P78555, Q8NFD1 | Gene names | ADARB1, ADAR2, DRADA2, RED1 | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific editase 1 (EC 3.5.-.-) (dsRNA adenosine deaminase) (RNA-editing deaminase 1) (RNA-editing enzyme 1). | |||||
|
EPHA4_MOUSE
|
||||||
| NC score | 0.006846 (rank : 81) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03137 | Gene names | Epha4, Sek, Sek1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (MPK-3). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.005450 (rank : 82) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.005243 (rank : 83) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
OXSR1_HUMAN
|
||||||
| NC score | 0.005149 (rank : 84) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95747, Q3LR53, Q7Z501, Q9UPQ1 | Gene names | OXSR1, KIAA1101, OSR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
EPHA4_HUMAN
|
||||||
| NC score | 0.004294 (rank : 85) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54764 | Gene names | EPHA4, HEK8, SEK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (Receptor protein-tyrosine kinase HEK8). | |||||
|
ZN295_HUMAN
|
||||||
| NC score | 0.002769 (rank : 86) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ3, Q6P4R0 | Gene names | ZNF295, KIAA1227, ZBTB21 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 295 (Zinc finger and BTB domain-containing protein 21). | |||||
|
ZIC5_HUMAN
|
||||||
| NC score | 0.001973 (rank : 87) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T25 | Gene names | ZIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 5 (Zinc finger protein of the cerebellum 5). | |||||
|
PRDM4_HUMAN
|
||||||
| NC score | 0.000428 (rank : 88) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 88 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKN5, Q9UFA6 | Gene names | PRDM4, PFM1 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 4 (PR domain-containing protein 4). | |||||