Please be patient as the page loads
|
CAP1_HUMAN
|
||||||
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |
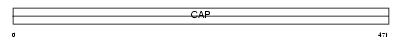
|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995962 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |
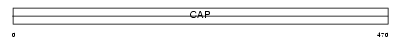
|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP2_HUMAN
|
||||||
| θ value | 1.41924e-154 (rank : 3) | NC score | 0.982151 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |
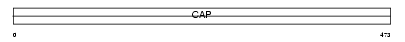
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CAP2_MOUSE
|
||||||
| θ value | 2.12077e-150 (rank : 4) | NC score | 0.984178 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |
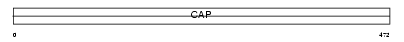
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.024739 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.015717 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 7) | NC score | 0.032499 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.010785 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
YTHD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.030638 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
MARE1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.026897 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15691 | Gene names | MAPRE1 | |||
|
Domain Architecture |
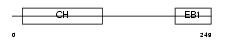
|
|||||
| Description | Microtubule-associated protein RP/EB family member 1 (APC-binding protein EB1) (End-binding protein 1) (EB1). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.027970 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.027336 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.046711 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.010431 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.017054 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
NOS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.013724 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.008770 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.021160 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.013813 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.005722 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.005875 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.016654 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.016696 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.002964 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
UBP8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.006038 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.017870 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.016970 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
HOMEZ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.011185 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.011742 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CAP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP1_MOUSE
|
||||||
| NC score | 0.995962 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP2_MOUSE
|
||||||
| NC score | 0.984178 (rank : 3) | θ value | 2.12077e-150 (rank : 4) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CAP2_HUMAN
|
||||||
| NC score | 0.982151 (rank : 4) | θ value | 1.41924e-154 (rank : 3) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.046711 (rank : 5) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.032499 (rank : 6) | θ value | 1.06291 (rank : 7) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
YTHD1_HUMAN
|
||||||
| NC score | 0.030638 (rank : 7) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.027970 (rank : 8) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.027336 (rank : 9) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MARE1_HUMAN
|
||||||
| NC score | 0.026897 (rank : 10) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15691 | Gene names | MAPRE1 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein RP/EB family member 1 (APC-binding protein EB1) (End-binding protein 1) (EB1). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.024739 (rank : 11) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.021160 (rank : 12) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.017870 (rank : 13) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.017054 (rank : 14) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.016970 (rank : 15) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.016696 (rank : 16) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.016654 (rank : 17) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.015717 (rank : 18) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.013813 (rank : 19) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
NOS3_HUMAN
|
||||||
| NC score | 0.013724 (rank : 20) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.011742 (rank : 21) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
HOMEZ_HUMAN
|
||||||
| NC score | 0.011185 (rank : 22) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.010785 (rank : 23) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.010431 (rank : 24) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.008770 (rank : 25) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
UBP8_MOUSE
|
||||||
| NC score | 0.006038 (rank : 26) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U87, Q80YP2, Q8R0D3, Q9EQU1, Q9WVP5 | Gene names | Usp8, Kiaa0055, Ubpy | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (mUBPy). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.005875 (rank : 27) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.005722 (rank : 28) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.002964 (rank : 29) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 29 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||