Please be patient as the page loads
|
CAP1_MOUSE
|
||||||
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |
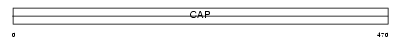
|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995962 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |
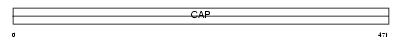
|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP2_HUMAN
|
||||||
| θ value | 3.16172e-154 (rank : 3) | NC score | 0.981527 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |
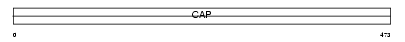
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CAP2_MOUSE
|
||||||
| θ value | 8.0589e-150 (rank : 4) | NC score | 0.982811 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |
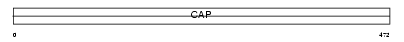
|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.015299 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.037637 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.025795 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.018501 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.022701 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.014295 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.017124 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.029905 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.033957 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
YTHD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.029188 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.026770 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.005736 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.009933 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
SF3B2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.045301 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.012762 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DACT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.059910 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.013734 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.020814 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.016028 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
ZN598_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.019666 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.004771 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ANKS3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.005383 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.016205 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.001106 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
HES2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.015537 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
MARE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.023279 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15691 | Gene names | MAPRE1 | |||
|
Domain Architecture |
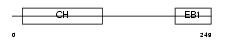
|
|||||
| Description | Microtubule-associated protein RP/EB family member 1 (APC-binding protein EB1) (End-binding protein 1) (EB1). | |||||
|
MOT8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.012714 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.018529 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.026362 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.014099 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.018211 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.010058 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MLE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.008626 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
MYL4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.009014 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.005520 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.007018 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
DOK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.008635 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |
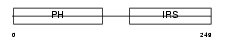
|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
GAB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.008836 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
NOL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.015089 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |
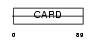
|
|||||
| Description | Nucleolar protein 3. | |||||
|
NOS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.011191 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.018699 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.012839 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
CAP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P40124 | Gene names | Cap1, Cap | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP1_HUMAN
|
||||||
| NC score | 0.995962 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01518, Q53HR7, Q5T0S1, Q6I9U6 | Gene names | CAP1, CAP | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 1 (CAP 1). | |||||
|
CAP2_MOUSE
|
||||||
| NC score | 0.982811 (rank : 3) | θ value | 8.0589e-150 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CYT6, Q80YU7, Q9D6L0 | Gene names | Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
CAP2_HUMAN
|
||||||
| NC score | 0.981527 (rank : 4) | θ value | 3.16172e-154 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40123, Q6IAY2 | Gene names | CAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Adenylyl cyclase-associated protein 2 (CAP 2). | |||||
|
DACT1_MOUSE
|
||||||
| NC score | 0.059910 (rank : 5) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4A3, Q80VG9, Q8BP49, Q9JK89 | Gene names | Dact1, Thyex3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (MDpr1) (Frodo) (Thymus-expressed novel gene 3 protein). | |||||
|
SF3B2_HUMAN
|
||||||
| NC score | 0.045301 (rank : 6) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13435 | Gene names | SF3B2, SAP145 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 2 (Spliceosome-associated protein 145) (SAP 145) (SF3b150) (Pre-mRNA-splicing factor SF3b 145 kDa subunit). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.037637 (rank : 7) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.033957 (rank : 8) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.029905 (rank : 9) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
YTHD1_HUMAN
|
||||||
| NC score | 0.029188 (rank : 10) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYJ9, Q8N3G5, Q8TBT1, Q96AN4, Q96S57, Q9BTI7, Q9NX79 | Gene names | YTHDF1, C20orf21 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain family protein 1 (Dermatomyositis associated with cancer putative autoantigen 1) (DACA-1). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.026770 (rank : 11) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.026362 (rank : 12) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.025795 (rank : 13) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MARE1_HUMAN
|
||||||
| NC score | 0.023279 (rank : 14) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15691 | Gene names | MAPRE1 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein RP/EB family member 1 (APC-binding protein EB1) (End-binding protein 1) (EB1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.022701 (rank : 15) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.020814 (rank : 16) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ZN598_HUMAN
|
||||||
| NC score | 0.019666 (rank : 17) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UK7, Q8IW49, Q8N3D9, Q96FG3, Q9H7J3 | Gene names | ZNF598 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 598. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.018699 (rank : 18) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.018529 (rank : 19) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.018501 (rank : 20) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.018211 (rank : 21) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.017124 (rank : 22) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.016205 (rank : 23) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.016028 (rank : 24) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.015537 (rank : 25) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.015299 (rank : 26) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NOL3_MOUSE
|
||||||
| NC score | 0.015089 (rank : 27) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D1X0 | Gene names | Nol3 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein 3. | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.014295 (rank : 28) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.014099 (rank : 29) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.013734 (rank : 30) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.012839 (rank : 31) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.012762 (rank : 32) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.012714 (rank : 33) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
NOS3_HUMAN
|
||||||
| NC score | 0.011191 (rank : 34) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29474, Q13662, Q14251, Q14434 | Gene names | NOS3 | |||
|
Domain Architecture |

|
|||||
| Description | Nitric-oxide synthase, endothelial (EC 1.14.13.39) (EC-NOS) (NOS type III) (NOSIII) (Endothelial NOS) (eNOS) (Constitutive NOS) (cNOS). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.010058 (rank : 35) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.009933 (rank : 36) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
MYL4_HUMAN
|
||||||
| NC score | 0.009014 (rank : 37) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12829, P11783 | Gene names | MYL4, MLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light polypeptide 4 (Myosin light chain 1, embryonic muscle/atrial isoform) (Myosin light chain alkali, GT-1 isoform). | |||||
|
GAB2_MOUSE
|
||||||
| NC score | 0.008836 (rank : 38) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1S8, Q9R1X3 | Gene names | Gab2 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 2 (GRB2-associated binder 2) (PH domain-containing adaptor molecule p97). | |||||
|
DOK1_HUMAN
|
||||||
| NC score | 0.008635 (rank : 39) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99704, O43204, Q9UHG6 | Gene names | DOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)) (pp62). | |||||
|
MLE1_MOUSE
|
||||||
| NC score | 0.008626 (rank : 40) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05977 | Gene names | Myl1, Mylf | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain 1, skeletal muscle isoform (MLC1F) (A1 catalytic) (Alkali myosin light chain 1). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.007018 (rank : 41) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.005736 (rank : 42) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.005520 (rank : 43) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
ANKS3_MOUSE
|
||||||
| NC score | 0.005383 (rank : 44) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZK6, Q6ZPF6, Q80X46 | Gene names | Anks3, Kiaa1977 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 3. | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.004771 (rank : 45) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.001106 (rank : 46) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||